3HYF
 
 | | Crystal structure of HIV-1 RNase H p15 with engineered E. coli loop and active site inhibitor | | 分子名称: | 2-(3,4-dichlorobenzyl)-5,6-dihydroxypyrimidine-4-carboxylic acid, ACETATE ION, GLYCEROL, ... | | 著者 | Lansdon, E.B, Kirschberg, T.A. | | 登録日 | 2009-06-22 | | 公開日 | 2009-10-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | RNase H active site inhibitors of human immunodeficiency virus type 1 reverse transcriptase: design, biochemical activity, and structural information.
J.Med.Chem., 52, 2009
|
|
1OVL
 
 | | Crystal Structure of Nurr1 LBD | | 分子名称: | BROMIDE ION, IODIDE ION, Orphan nuclear receptor NURR1 (MSE 414, ... | | 著者 | Wang, Z, Liu, J, Walker, N. | | 登録日 | 2003-03-26 | | 公開日 | 2003-06-03 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure and Function of Nurr1 identifies a Class of Ligand-Independent Nuclear Receptors
Nature, 423, 2003
|
|
8I88
 
 | | Cryo-EM structure of TIR-APAZ/Ago-gRNA complex | | 分子名称: | Piwi domain-containing protein, RNA (5'-R(P*GP*A)-3'), TIR domain-containing protein | | 著者 | Zhang, H, Li, Z, Yu, G.M, Li, X.Z, Wang, X.S. | | 登録日 | 2023-02-03 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity.
Cell Res., 33, 2023
|
|
8I87
 
 | | Cryo-EM structure of TIR-APAZ/Ago-gRNA-DNA complex | | 分子名称: | DNA (5'-D(P*TP*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*A)-3'), MAGNESIUM ION, Piwi domain-containing protein, ... | | 著者 | Zhang, H, Deng, Z.Q, Yu, G.M, Li, X.Z, Wang, X.S. | | 登録日 | 2023-02-03 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-09-13 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity.
Cell Res., 33, 2023
|
|
8Y8D
 
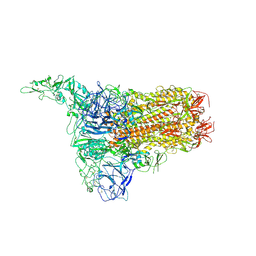 | | Structure of HCoV-HKU1C spike in the inactive-1up conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8I
 
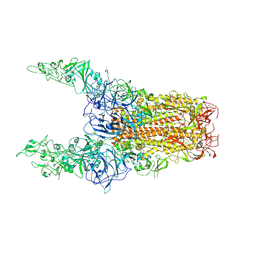 | | Structure of HCoV-HKU1C spike in the glycan-activated-3up conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8C
 
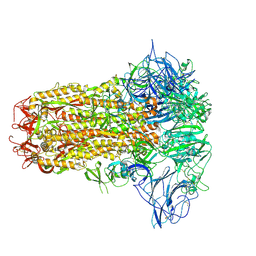 | | Structure of HCoV-HKU1C spike in the inactive-closed conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8E
 
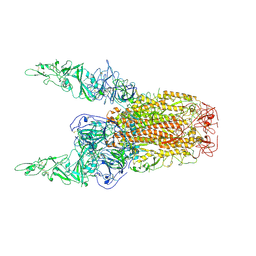 | | Structure of HCoV-HKU1C spike in the inactive-2up conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8G
 
 | | Structure of HCoV-HKU1C spike in the glycan-activated-1up conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8H
 
 | | Structure of HCoV-HKU1C spike in the glycan-activated-2up conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8J
 
 | | Local structure of HCoV-HKU1C spike in complex with glycan | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.57 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8F
 
 | | Structure of HCoV-HKU1C spike in the glycan-activated-closed conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8HSB
 
 | |
2EXX
 
 | | Crystal structure of HSCARG from Homo sapiens in complex with NADP | | 分子名称: | GLYCEROL, HSCARG protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Dai, X, Chen, Q, Yao, D, Liang, Y, Dong, Y, Gu, X, Zheng, X, Luo, M. | | 登録日 | 2005-11-09 | | 公開日 | 2006-11-21 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Restructuring of the dinucleotide-binding fold in an NADP(H) sensor protein.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
6TD5
 
 | | Leishmania tarentolae proteasome 20S subunit complexed with LXE408 and bortezomib | | 分子名称: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, Proteasome endopeptidase complex, Proteasome subunit alpha type, ... | | 著者 | Srinivas, H. | | 登録日 | 2019-11-07 | | 公開日 | 2020-08-26 | | 最終更新日 | 2020-10-21 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Discovery and Characterization of Clinical Candidate LXE408 as a Kinetoplastid-Selective Proteasome Inhibitor for the Treatment of Leishmaniases.
J.Med.Chem., 63, 2020
|
|
6TCZ
 
 | | Leishmania tarentolae proteasome 20S subunit complexed with LXE408 | | 分子名称: | Proteasome endopeptidase complex, Proteasome subunit alpha type, Proteasome subunit beta, ... | | 著者 | Srinivas, H. | | 登録日 | 2019-11-07 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Discovery and Characterization of Clinical Candidate LXE408 as a Kinetoplastid-Selective Proteasome Inhibitor for the Treatment of Leishmaniases.
J.Med.Chem., 63, 2020
|
|
1B4P
 
 | | CRYSTAL STRUCTURES OF CLASS MU CHIMERIC GST ISOENZYMES M1-2 AND M2-1 | | 分子名称: | L-gamma-glutamyl-S-[(9S,10S)-10-hydroxy-9,10-dihydrophenanthren-9-yl]-L-cysteinylglycine, PROTEIN (GLUTATHIONE S-TRANSFERASE), SULFATE ION | | 著者 | Xiao, G, Chen, J, Armstrong, R.N, Gilliland, G.L. | | 登録日 | 1998-12-26 | | 公開日 | 2003-07-08 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structures of Class MU Chimeric GST Isoenzymes M1-2 and M2-1
To be Published
|
|
8XEP
 
 | | Crystal structure of a Legionella pneumophila type IV effector in complex with ubiquitin | | 分子名称: | SULFATE ION, Type IV effector MavL, Ubiquitin | | 著者 | Tan, J.X, Wang, X.F, Zhou, Y, Zhu, Y.Q. | | 登録日 | 2023-12-12 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Legionella effector LnaB is a phosphoryl AMPylase that impairs phosphosignalling.
Nature, 631, 2024
|
|
5IVC
 
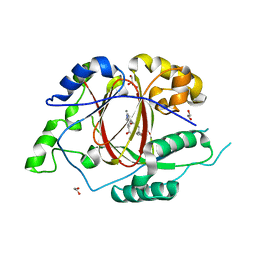 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N3 (4'-[(2-phenylethyl)carbamoyl][2,2'-bipyridine]-4-carboxylic acid) | | 分子名称: | 1,2-ETHANEDIOL, 4'-[(2-phenylethyl)carbamoyl][2,2'-bipyridine]-4-carboxylic acid, GLYCEROL, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2016-03-20 | | 公開日 | 2016-07-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.573 Å) | | 主引用文献 | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
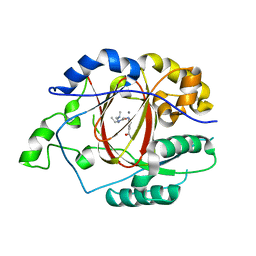
5IWF
 
 | |
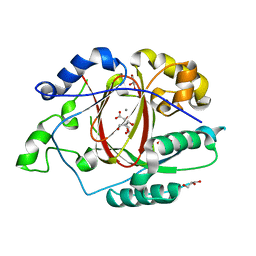
5IVB
 
 | |
5IVF
 
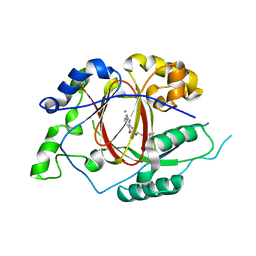 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N10 8-(1-methyl-1H-imidazol-4-yl)-2-(4,4,4-trifluorobutoxy)pyrido[3,4-d]pyrimidin-4-ol | | 分子名称: | 8-(1-methyl-1H-imidazol-4-yl)-2-(4,4,4-trifluorobutoxy)pyrido[3,4-d]pyrimidin-4-ol, Lysine-specific demethylase 5A, MANGANESE (II) ION | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2016-03-20 | | 公開日 | 2016-07-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.683 Å) | | 主引用文献 | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5IVV
 
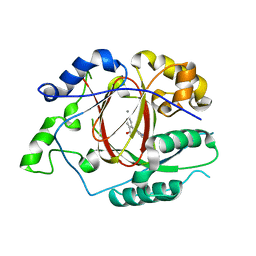 | |
5IVJ
 
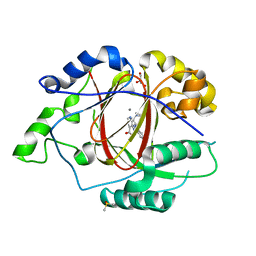 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N11 [3-({1-[2-(4,4-difluoropiperidin-1-yl)ethyl]-5-fluoro-1H-indazol-3-yl}amino)pyridine-4-carboxylic acid] | | 分子名称: | 3-({1-[2-(4,4-difluoropiperidin-1-yl)ethyl]-5-fluoro-1H-indazol-3-yl}amino)pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2016-03-21 | | 公開日 | 2016-07-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.569 Å) | | 主引用文献 | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5QJ2
 
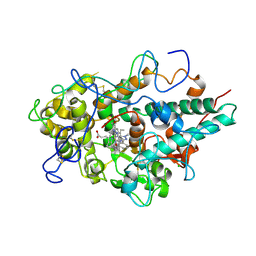 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) OMPLEX WITH COMPOUND-20 AKA 7-((3-(1-METHYL-1H-PYRAZOL-3- YL)BENZYL)OXY)- 1H-[1,2,3]TRIAZOLO[4,5-B]PYRIDIN-5-AMINE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-{[3-(1-methyl-1H-pyrazol-3-yl)phenyl]methoxy}-1H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, CALCIUM ION, ... | | 著者 | Khan, J.A. | | 登録日 | 2018-09-26 | | 公開日 | 2019-02-06 | | 最終更新日 | 2021-05-12 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | Potent Triazolopyridine Myeloperoxidase Inhibitors.
ACS Med Chem Lett, 9, 2018
|
|
