7XU3
 
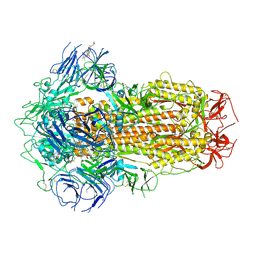 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU6
 
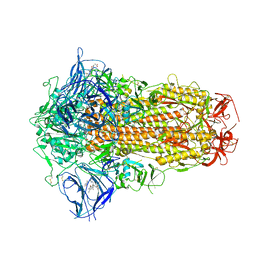 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), incubated in Low pH after 40-Day Storage in PBS, Locked-2 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
5TY7
 
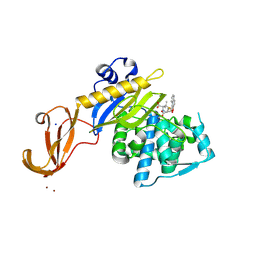 | | Crystal structure of wild-type S. aureus penicillin binding protein 4 (PBP4) in complex with nafcillin | | 分子名称: | (2R,4S)-2-[(1R)-1-{[(2-ethoxynaphthalen-1-yl)carbonyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 4, SODIUM ION, ... | | 著者 | Alexander, J.A.N, Strynadka, N.C.J. | | 登録日 | 2016-11-18 | | 公開日 | 2018-06-13 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.894 Å) | | 主引用文献 | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
7KCX
 
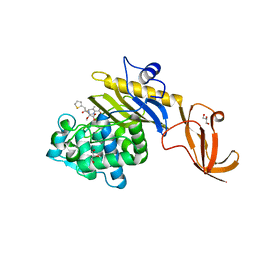 | | Crystal structure of S. aureus penicillin-binding protein 4 (PBP4) mutant (R200L) in complex with cefoxitin | | 分子名称: | (2R)-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4, ... | | 著者 | Alexander, J.A.N, Strynadka, N.C.J. | | 登録日 | 2020-10-07 | | 公開日 | 2021-06-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | PBP4-mediated beta-lactam resistance among clinical strains of Staphylococcus aureus.
J.Antimicrob.Chemother., 76, 2021
|
|
7KCV
 
 | |
7KCY
 
 | | Crystal structure of S. aureus penicillin-binding protein 4 (PBP4) with cefoxitin | | 分子名称: | (2R)-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4, ... | | 著者 | Alexander, J.A.N, Strynadka, N.C.J. | | 登録日 | 2020-10-07 | | 公開日 | 2021-06-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | PBP4-mediated beta-lactam resistance among clinical strains of Staphylococcus aureus.
J.Antimicrob.Chemother., 76, 2021
|
|
7JGX
 
 | |
7JGY
 
 | |
7JHF
 
 | |
5W4C
 
 | | Crystal structure of thioredoxin reductase from Cryptococcus neoformans in complex with FAD (FO conformation) | | 分子名称: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Bravo-Chaucanes, C.P, Valadares, N.F, Felipe, M.S.S, Barbosa, J.A.R.G. | | 登録日 | 2017-06-09 | | 公開日 | 2018-06-13 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.255 Å) | | 主引用文献 | Crystal structure of thioredoxin reductase from Cryptococcus neoformans in complex with FAD (FO conformation)
To be published
|
|
6FPX
 
 | | Structure of S. pombe Mmi1 in complex with 11-mer RNA | | 分子名称: | GLYCEROL, RNA (5'-R(P*UP*UP*UP*AP*AP*AP*CP*CP*UP*A)-3'), YTH domain-containing protein mmi1 | | 著者 | Stowell, J.A.W, Hill, C.H, Yu, M, Wagstaff, J.L, McLaughlin, S.H, Freund, S.M.V, Passmore, L.A. | | 登録日 | 2018-02-12 | | 公開日 | 2018-05-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | A low-complexity region in the YTH domain protein Mmi1 enhances RNA binding.
J. Biol. Chem., 293, 2018
|
|
1E95
 
 | | Solution structure of the pseudoknot of SRV-1 RNA, involved in ribosomal frameshifting | | 分子名称: | RNA (5'-(*GP*CP*GP*GP*CP*CP*AP*GP*CP*UP*CP* CP*AP*GP*GP*CP*CP*GP*CP*CP*AP*AP*AP*CP* AP*AP*UP*AP*UP*GP*GP*AP*GP*CP*AP*C)-3') | | 著者 | Michiels, P.J.A, Versleyen, A, Pleij, C.W.A, Hilbers, C.W, Heus, H.A. | | 登録日 | 2000-10-09 | | 公開日 | 2001-08-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the Pseudoknot of Srv-1 RNA, Involved in Ribosomal Frameshifting
J.Mol.Biol., 310, 2001
|
|
2DQ5
 
 | | solution structure of the Mid1 B Box2 Chc(D/C)C2H2 Zinc-Binding Domain: insights into an evolutionary conserved ring fold | | 分子名称: | Midline-1, ZINC ION | | 著者 | Massiah, M.A, Matts, J.A.B, Short, K.M, Simmons, B.N, Singireddy, S, Zou, J, Cox, T.C. | | 登録日 | 2006-05-20 | | 公開日 | 2007-04-03 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the MID1 B-box2 CHC(D/C)C(2)H(2) Zinc-binding Domain: Insights into an Evolutionarily Conserved RING Fold
J.Mol.Biol., 369, 2007
|
|
1F6K
 
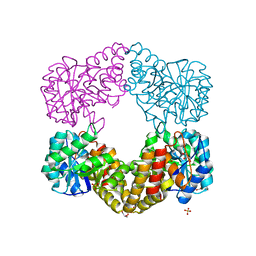 | | CRYSTAL STRUCTURE ANALYSIS OF N-ACETYLNEURAMINATE LYASE FROM HAEMOPHILUS INFLUENZAE: CRYSTAL FORM II | | 分子名称: | GLYCEROL, N-ACETYLNEURAMINATE LYASE, SULFATE ION | | 著者 | Barbosa, J.A.R.G, Smith, B.J, DeGori, R, Lawrence, M.C. | | 登録日 | 2000-06-21 | | 公開日 | 2000-12-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Active site modulation in the N-acetylneuraminate lyase sub-family as revealed by the structure of the inhibitor-complexed Haemophilus influenzae enzyme.
J.Mol.Biol., 303, 2000
|
|
1F5Z
 
 | |
6DGE
 
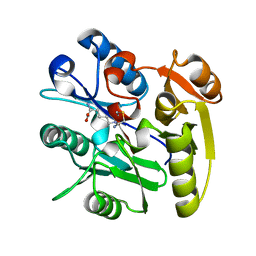 | | Crystal structure of the dimethylarginine dimethylaminohydrolase adduct with N5-(1-imino-2-chloroethyl)-L-lysine | | 分子名称: | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1, N~6~-[(1E)-2-chloroethanimidoyl]-L-lysine | | 著者 | Monzingo, A.F, Burstein-Teitelbaum, G, Er, J.A.V, Tuley, A, Fast, W. | | 登録日 | 2018-05-17 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Dissection, Optimization, and Structural Analysis of a Covalent Irreversible DDAH1 Inhibitor.
Biochemistry, 57, 2018
|
|
6DZ8
 
 | |
6EHM
 
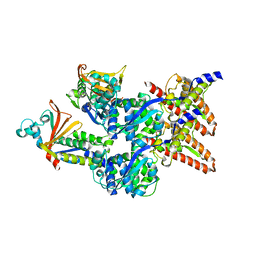 | | Model of the Ebola virus nucleocapsid subunit from recombinant virus-like particles | | 分子名称: | Membrane-associated protein VP24, Nucleoprotein | | 著者 | Wan, W, Kolesnikova, L, Clarke, M, Koehler, A, Noda, T, Becker, S, Briggs, J.A.G. | | 登録日 | 2017-09-13 | | 公開日 | 2017-11-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (7.3 Å) | | 主引用文献 | Structure and assembly of the Ebola virus nucleocapsid.
Nature, 551, 2017
|
|
6EI0
 
 | | Cytosolic copper storage protein Csp from Streptomyces lividans: apo form | | 分子名称: | Cytosolic copper storage protein (Ccsp), GLYCEROL, SULFATE ION, ... | | 著者 | Straw, M.L, Chaplin, A.K, Hough, M.A, Worrall, J.A.R. | | 登録日 | 2017-09-15 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | A cytosolic copper storage protein provides a second level of copper tolerance in Streptomyces lividans.
Metallomics, 10, 2018
|
|
6EAV
 
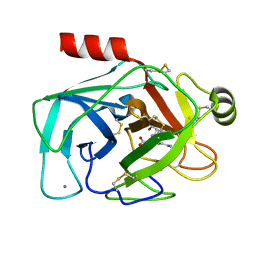 | |
1GKK
 
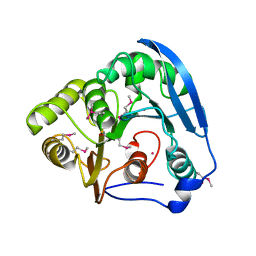 | | Feruloyl esterase domain of XynY from clostridium thermocellum | | 分子名称: | CADMIUM ION, ENDO-1,4-BETA-XYLANASE Y, GLYCEROL | | 著者 | Prates, J.A.M, Tarbouriech, N, Charnock, S.J, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J. | | 登録日 | 2001-08-15 | | 公開日 | 2001-12-13 | | 最終更新日 | 2011-09-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The Structure of the Feruloyl Esterase Module of Xylanase 10B from Clostridium Thermocellum Provides Insights Into Substrate Recognition
Structure, 9, 2001
|
|
6EAW
 
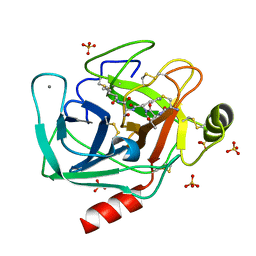 | | Crystallographic structure of the cyclic heptapeptide derived from the BTCI inhibitor bound to beta-trypsin in space group P 21 21 21 | | 分子名称: | CALCIUM ION, CYS-THR-LYS-SER-ILE, Cationic trypsin, ... | | 著者 | Fernandes, J.C, Valadares, N.F, Freitas, S.M, Barbosa, J.A.R.G. | | 登録日 | 2018-08-03 | | 公開日 | 2019-08-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.289 Å) | | 主引用文献 | Crystallographic structure of the cyclic heptapeptide derived from the BTCI inhibitor bound to beta-trypsin in space group P 21 21 21
To Be Published
|
|
6EHL
 
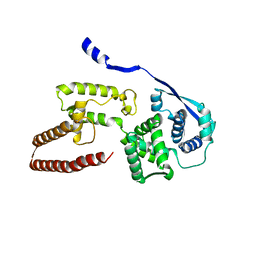 | | Model of the Ebola virus nucleoprotein in recombinant nucleocapsid-like assemblies | | 分子名称: | Nucleoprotein | | 著者 | Wan, W, Kolesnikova, L, Clarke, M, Koehler, A, Noda, T, Becker, S, Briggs, J.A.G. | | 登録日 | 2017-09-13 | | 公開日 | 2017-11-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (6.6 Å) | | 主引用文献 | Structure and assembly of the Ebola virus nucleocapsid.
Nature, 551, 2017
|
|
6EAX
 
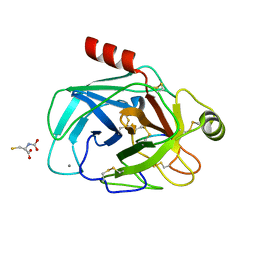 | | Crystallographic structure of the cyclic hexapeptide derived from the BTCI inhibitor bound to beta-trypsin in space group P 21 21 21 | | 分子名称: | CALCIUM ION, CYS-THR-LYS-SER-ILE-CYS, Cationic trypsin, ... | | 著者 | Fernandes, J.C, Valadares, N.F, Freitas, S.M, Barbosa, J.A.R.G. | | 登録日 | 2018-08-03 | | 公開日 | 2019-08-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.189 Å) | | 主引用文献 | Crystallographic structure of the cyclic hexapeptide derived from the BTCI inhibitor bound to beta-trypsin in space group P 21 21 21
To Be Published
|
|
6H7W
 
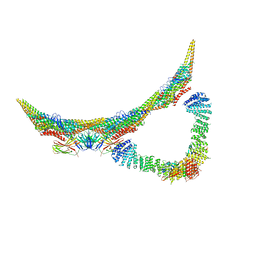 | | Model of retromer-Vps5 complex assembled on membrane. | | 分子名称: | Putative vacuolar protein sorting-associated protein, Vacuolar protein sorting-associated protein 26-like protein, Vacuolar protein sorting-associated protein 29, ... | | 著者 | Kovtun, O, Leneva, N, Ariotti, N, Rohan, T.S, Owen, D.J, Briggs, J.A.G, Collins, B.M. | | 登録日 | 2018-07-31 | | 公開日 | 2018-09-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (11.4 Å) | | 主引用文献 | Structure of the membrane-assembled retromer coat determined by cryo-electron tomography.
Nature, 561, 2018
|
|
