3GDU
 
 | |
3G1B
 
 | | The structure of the M53A mutant of Caulobacter crescentus clpS protease adaptor protein in complex with WLFVQRDSKE peptide | | 分子名称: | 10-residue peptide, ATP-dependent Clp protease adapter protein clpS, MAGNESIUM ION | | 著者 | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | 登録日 | 2009-01-29 | | 公開日 | 2009-04-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.448 Å) | | 主引用文献 | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GDV
 
 | |
3GES
 
 | |
3G19
 
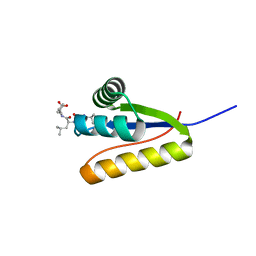 | | The structure of the Caulobacter crescentus clpS protease adaptor protein in complex with LLL tripeptide | | 分子名称: | ATP-dependent Clp protease adapter protein clpS, LLL tripeptide | | 著者 | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | 登録日 | 2009-01-29 | | 公開日 | 2009-04-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.849 Å) | | 主引用文献 | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GER
 
 | |
3G3P
 
 | |
3GAO
 
 | |
3GCO
 
 | |
3GDS
 
 | |
3EQ2
 
 | |
3GQ0
 
 | |
3GOT
 
 | | Guanine riboswitch C74U mutant bound to 2-fluoroadenine. | | 分子名称: | 2-fluoroadenine, ACETATE ION, COBALT HEXAMMINE(III), ... | | 著者 | Gilbert, S.D, Reyes, F.E, Batey, R.T. | | 登録日 | 2009-03-20 | | 公開日 | 2009-06-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Adaptive ligand binding by the purine riboswitch in the recognition of Guanine and adenine analogs
Structure, 17, 2009
|
|
3GW1
 
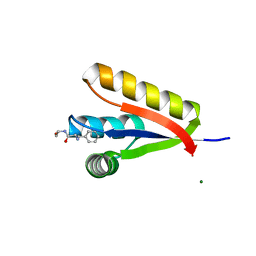 | | The structure of the Caulobacter crescentus CLPs protease adaptor protein in complex with FGG tripeptide | | 分子名称: | ATP-dependent Clp protease adapter protein ClpS, FGG peptide, MAGNESIUM ION | | 著者 | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | 登録日 | 2009-03-31 | | 公開日 | 2009-05-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3DS7
 
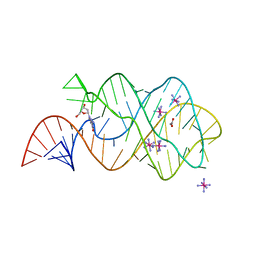 | |
3GQ1
 
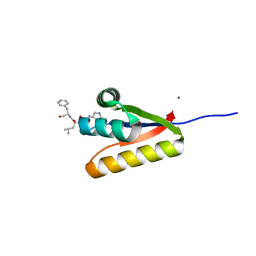 | | The structure of the caulobacter crescentus clpS protease adaptor protein in complex with a WLFVQRDSKE decapeptide | | 分子名称: | ATP-dependent Clp protease adapter protein clpS, MAGNESIUM ION, WLFVQRDSKE peptide | | 著者 | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | 登録日 | 2009-03-23 | | 公開日 | 2009-05-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.496 Å) | | 主引用文献 | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3FO4
 
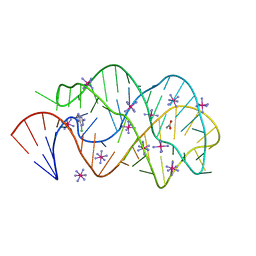 | |
3F79
 
 | |
3GX2
 
 | |
3GQX
 
 | |
3GX5
 
 | |
3GQU
 
 | |
3GX6
 
 | |
3DD1
 
 | | Crystal structure of glycogen phophorylase complexed with an anthranilimide based inhibitor GSK254 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-cyclohexyl-N-[(3-{[(2,4,6-trimethylphenyl)carbamoyl]amino}naphthalen-2-yl)carbonyl]-D-alanine, ... | | 著者 | Nolte, R.T. | | 登録日 | 2008-06-04 | | 公開日 | 2009-04-21 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Anthranilimide based glycogen phosphorylase inhibitors for the treatment of type 2 diabetes. Part 3: X-ray crystallographic characterization, core and urea optimization and in vivo efficacy.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3DDS
 
 | |
