7L0M
 
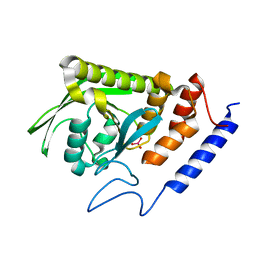 | | Vanadate-bound YopH G352T | | 分子名称: | Protein-tyrosine-phosphatase, VANADATE ION | | 著者 | Shen, R.D, Hengge, A.C, Johnson, S.J. | | 登録日 | 2020-12-11 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Single Residue on the WPD-Loop Affects the pH Dependency of Catalysis in Protein Tyrosine Phosphatases.
Jacs Au, 1, 2021
|
|
7L0H
 
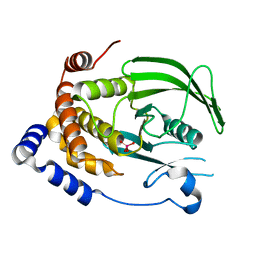 | | Vanadate-bound PTP1B T177G | | 分子名称: | Tyrosine-protein phosphatase non-receptor type 1, VANADATE ION | | 著者 | Shen, R.D, Hengge, A.C, Johnson, S.J. | | 登録日 | 2020-12-11 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Single Residue on the WPD-Loop Affects the pH Dependency of Catalysis in Protein Tyrosine Phosphatases.
Jacs Au, 1, 2021
|
|
4UHU
 
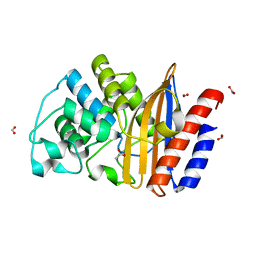 | | W229D mutant of the last common ancestor of Gram-negative bacteria (GNCA) beta-lactamase class A | | 分子名称: | ACETATE ION, FORMIC ACID, GNCA LACTAMASE W229D | | 著者 | Gavira, J.A, Risso, V.A, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M. | | 登録日 | 2015-03-25 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.305 Å) | | 主引用文献 | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
5FQQ
 
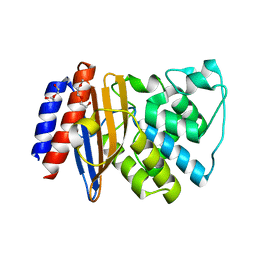 | | Last common ancestor of Gram-negative bacteria (GNCA4) beta-lactamase class A | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, DI(HYDROXYETHYL)ETHER, GNCA4 LACTAMASE | | 著者 | Gavira, J.A, Martinez-Rodriguez, S, Risso, V.A, Sanchez-Ruiz, J.M. | | 登録日 | 2015-12-14 | | 公開日 | 2016-12-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
5FQM
 
 | | Last common ancestor of Gram Negative Bacteria (GNCA) Class A beta- lactamase | | 分子名称: | GLYCEROL, GNCA BETA LACTAMASE, SULFATE ION | | 著者 | Martinez Rodriguez, S, Gavira, J.A, Risso, V.A, Sanchez Ruiz, J.M. | | 登録日 | 2015-12-12 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
5FQK
 
 | | W229D and F290W mutant of the last common ancestor of Gram-negative bacteria (GNCA4) beta-lactamase class A bound to 5(6)-nitrobenzotriazole (TS-analog) | | 分子名称: | 6-NITROBENZOTRIAZOLE, GNCA4 LACTAMASE W229D AND F290W | | 著者 | Gavira, J.A, Risso, V.A, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M. | | 登録日 | 2015-12-11 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.767 Å) | | 主引用文献 | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
4CYR
 
 | | G4 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa | | 分子名称: | ARYLSULFATASE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | 登録日 | 2014-04-14 | | 公開日 | 2015-04-29 | | 最終更新日 | 2019-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5FQJ
 
 | | W229D mutant of the last common ancestor of Gram-negative bacteria (GNCA) beta-lactamase bound to 5(6)-nitrobenzotriazole (TS-analog) | | 分子名称: | 6-NITROBENZOTRIAZOLE, GNCA LACTAMASE W229D | | 著者 | Gavira, J.A, Martinez-Rodriguez, S, Risso, V.A, Sanchez-Ruiz, J.M. | | 登録日 | 2015-12-11 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.271 Å) | | 主引用文献 | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
5FQI
 
 | | W229D and F290W mutant of the last common ancestor of Gram-negative bacteria (GNCA4) beta-lactamase class A | | 分子名称: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Gavira, J.A, Risso, V.A, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M. | | 登録日 | 2015-12-11 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
4CYS
 
 | | G6 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa, in complex with Phenylphosphonic acid | | 分子名称: | AMMONIUM ION, ARYLSULFATASE, CALCIUM ION, ... | | 著者 | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | 登録日 | 2014-04-14 | | 公開日 | 2015-04-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4CXU
 
 | | G4 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa, in complex with 3-Br-Phenolphenylphosphonate | | 分子名称: | 3-bromophenyl hydrogen (S)-phenylphosphonate, ARYLSULFATASE, CALCIUM ION | | 著者 | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | 登録日 | 2014-04-08 | | 公開日 | 2015-04-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4CXS
 
 | | G4 mutant of PAS, arylsulfatase from Pseudomonas aeruginosa, in complex with Phenylphosphonic acid | | 分子名称: | ARYLSULFATASE, CALCIUM ION, SULFATE ION, ... | | 著者 | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | 登録日 | 2014-04-08 | | 公開日 | 2015-05-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4CXK
 
 | | G9 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa | | 分子名称: | ARYLSULFATASE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | 登録日 | 2014-04-07 | | 公開日 | 2015-04-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C2C
 
 | | The molecular basis for the functional evolution of an organophosphate hydrolysing enzyme | | 分子名称: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ZINC ION, ... | | 著者 | Hong, N.-S, Jackson, C.J, Carr, P.D, Tokuriki, N, Baier, F, Yang, G. | | 登録日 | 2018-01-08 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.597 Å) | | 主引用文献 | Higher-order epistasis shapes the fitness landscape of a xenobiotic-degrading enzyme.
Nat.Chem.Biol., 15, 2019
|
|
8B67
 
 | |
8B6K
 
 | |
8B7E
 
 | |
8B79
 
 | |
8B77
 
 | |
8B76
 
 | |
6XEA
 
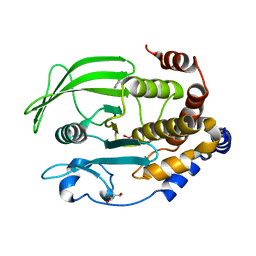 | | Crystal Structure of the PTP1B YopH WPD loop Chimera 3 bound to vanadate | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZAMIDINE, MAGNESIUM ION, ... | | 著者 | Olsen, K.J, Shen, R, Johnson, S.J, Hengge, A.C. | | 登録日 | 2020-06-12 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.549 Å) | | 主引用文献 | Insights into the importance of WPD-loop sequence for activity and structure in protein tyrosine phosphatases.
Chem Sci, 13, 2022
|
|
6XEE
 
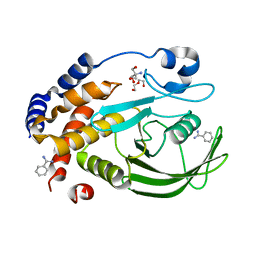 | | Crystal Structure of the PTP1B YopH WPD loop Chimera 4 apo form | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZAMIDINE, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Olsen, K.J, Shen, R, Johnson, S.J, Hengge, A.C. | | 登録日 | 2020-06-12 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Insights into the importance of WPD-loop sequence for activity and structure in protein tyrosine phosphatases.
Chem Sci, 13, 2022
|
|
6XE8
 
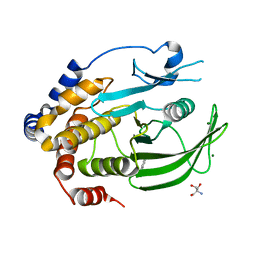 | | Crystal Structure of the PTP1B YopH WPD loop Chimera 3 apo form | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZAMIDINE, MAGNESIUM ION, ... | | 著者 | Olsen, K.J, Shen, R, Johnson, S.J, Hengge, A.C. | | 登録日 | 2020-06-12 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.952 Å) | | 主引用文献 | Insights into the importance of WPD-loop sequence for activity and structure in protein tyrosine phosphatases.
Chem Sci, 13, 2022
|
|
6XED
 
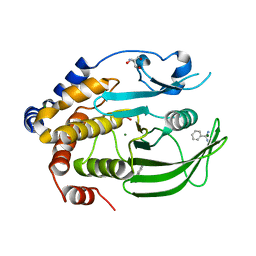 | | Crystal Structure of the PTP1B YopH WPD loop Chimera 3 bound to tungstate | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZAMIDINE, MAGNESIUM ION, ... | | 著者 | Olsen, K.J, Shen, R, Johnson, S.J, Hengge, A.C. | | 登録日 | 2020-06-12 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.795 Å) | | 主引用文献 | Insights into the importance of WPD-loop sequence for activity and structure in protein tyrosine phosphatases.
Chem Sci, 13, 2022
|
|
6XEF
 
 | | Crystal structure of the PTP1B YopH WPD loop Chimera 4 bound to vanadate | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZAMIDINE, MAGNESIUM ION, ... | | 著者 | Olsen, K.J, Shen, R, Johnson, S.J, Hengge, A.C. | | 登録日 | 2020-06-12 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.048 Å) | | 主引用文献 | Insights into the importance of WPD-loop sequence for activity and structure in protein tyrosine phosphatases.
Chem Sci, 13, 2022
|
|
