2O7X
 
 | | Duplex DNA containing an abasic site with an opposite G (beta anomer) in 5'-G_AC-3' (10 structure ensemble and averaged structure) | | 分子名称: | 5'-D(*CP*CP*AP*AP*AP*GP*(AAB)P*AP*CP*CP*GP*GP*G)-3', 5'-D(*CP*CP*CP*GP*GP*TP*GP*CP*TP*TP*TP*GP*G)-3' | | 著者 | Chen, J, Dupradeau, F.Y, Case, D.A, Turner, C.J, Stubbe, J. | | 登録日 | 2006-12-11 | | 公開日 | 2007-11-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | DNA oligonucleotides with A, T, G or C opposite an abasic site: structure and dynamics.
Nucleic Acids Res., 36, 2008
|
|
2O7Y
 
 | | Duplex DNA containing an abasic site with an opposite T (alpha anomer) in 5'-G_AC-3' (10 structure ensemble and averaged structure) | | 分子名称: | 5'-D(*CP*CP*AP*AP*AP*GP*(ORP)P*AP*CP*CP*GP*GP*G)-3', 5'-D(*CP*CP*CP*GP*GP*TP*TP*CP*TP*TP*TP*GP*G)-3' | | 著者 | Dupradeau, F.Y, Case, D.A, Turner, C.J, Stubbe, J. | | 登録日 | 2006-12-11 | | 公開日 | 2007-11-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | DNA oligonucleotides with A, T, G or C opposite an abasic site: structure and dynamics.
Nucleic Acids Res., 36, 2008
|
|
2O7W
 
 | | Duplex DNA containing an abasic site with an opposite G (alpha anomer) in 5'-G_AC-3' (10 structure ensemble and averaged structure) | | 分子名称: | 5'-D(*CP*CP*AP*AP*AP*GP*(ORP)P*AP*CP*CP*GP*GP*G)-3', 5'-D(*CP*CP*CP*GP*GP*TP*GP*CP*TP*TP*TP*GP*G)-3' | | 著者 | Chen, J, Dupradeau, F.Y, Case, D.A, Turner, C.J, Stubbe, J. | | 登録日 | 2006-12-11 | | 公開日 | 2007-11-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | DNA oligonucleotides with A, T, G or C opposite an abasic site: structure and dynamics.
Nucleic Acids Res., 36, 2008
|
|
2LEF
 
 | | LEF1 HMG DOMAIN (FROM MOUSE), COMPLEXED WITH DNA (15BP), NMR, 12 STRUCTURES | | 分子名称: | DNA (5'-D(*CP*AP*CP*CP*CP*TP*TP*TP*GP*AP*AP*GP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*CP*TP*TP*CP*AP*AP*AP*GP*GP*GP*TP*G)-3'), PROTEIN (LYMPHOID ENHANCER-BINDING FACTOR) | | 著者 | Li, X, Love, J.J, Case, D.A, Wright, P.E. | | 登録日 | 1998-10-13 | | 公開日 | 1998-10-21 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for DNA bending by the architectural transcription factor LEF-1.
Nature, 376, 1995
|
|
2NC6
 
 | | Solution Structure of N-L-idosylated Pin1 WW Domain | | 分子名称: | Pin1 WW Domain, beta-L-idopyranose | | 著者 | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | 登録日 | 2016-03-20 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
2NC3
 
 | | Solution Structure of N-Allosylated Pin1 WW Domain | | 分子名称: | Pin1 WW Domain, beta-D-allopyranose | | 著者 | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | 登録日 | 2016-03-20 | | 公開日 | 2016-06-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
2NC4
 
 | | Solution Structure of N-Galactosylated Pin1 WW Domain | | 分子名称: | Pin1 WW Domain, beta-D-galactopyranose | | 著者 | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | 登録日 | 2016-03-20 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
2NC5
 
 | | Solution Structure of N-Xylosylated Pin1 WW Domain | | 分子名称: | Pin1 WW Domain, beta-D-xylopyranose | | 著者 | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | 登録日 | 2016-03-20 | | 公開日 | 2016-06-08 | | 最終更新日 | 2020-07-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
2KBU
 
 | |
8DZ7
 
 | |
8DYZ
 
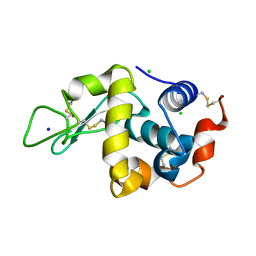 | |
6VVJ
 
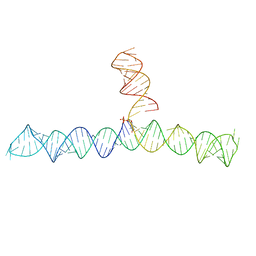 | | Cap1G-TPUA | | 分子名称: | RNA (130-MER) | | 著者 | Summers, M.F, Brown, J.D. | | 登録日 | 2020-02-18 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for transcriptional start site control of HIV-1 RNA fate.
Science, 368, 2020
|
|
6VU1
 
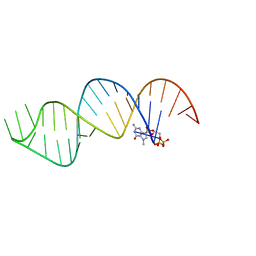 | |
1A03
 
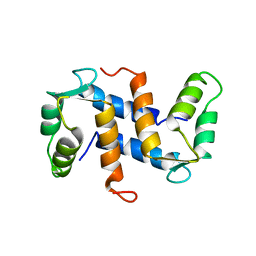 | | THE THREE-DIMENSIONAL STRUCTURE OF CA2+-BOUND CALCYCLIN: IMPLICATIONS FOR CA2+-SIGNAL TRANSDUCTION BY S100 PROTEINS, NMR, 20 STRUCTURES | | 分子名称: | CALCYCLIN (RABBIT, CA2+) | | 著者 | Sastry, M, Ketchem, R.R, Crescenzi, O, Weber, C, Lubienski, M.J, Hidaka, H, Chazin, W.J. | | 登録日 | 1997-12-08 | | 公開日 | 1999-03-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The three-dimensional structure of Ca(2+)-bound calcyclin: implications for Ca(2+)-signal transduction by S100 proteins.
Structure, 6, 1998
|
|
6BG9
 
 | |
6WXI
 
 | | Colicin E1 fragment in nanodisc-embedded TolC | | 分子名称: | Outer membrane protein TolC | | 著者 | Kaelber, J.T, Budiardjo, S.J, Firlar, E, Ikujuni, A.P, Slusky, J.S.G. | | 登録日 | 2020-05-10 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (2.84 Å) | | 主引用文献 | Colicin E1 opens its hinge to plug TolC.
Elife, 11, 2022
|
|
6O2H
 
 | |
1CUR
 
 | | REDUCED RUSTICYANIN, NMR | | 分子名称: | COPPER (II) ION, CU(I) RUSTICYANIN | | 著者 | Botuyan, M.V, Dyson, H.J. | | 登録日 | 1996-04-19 | | 公開日 | 1996-11-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of Cu(I) rusticyanin from Thiobacillus ferrooxidans: structural basis for the extreme acid stability and redox potential.
J.Mol.Biol., 263, 1996
|
|
1A01
 
 | | HEMOGLOBIN (VAL BETA1 MET, TRP BETA37 ALA) MUTANT | | 分子名称: | HEMOGLOBIN (ALPHA CHAIN), HEMOGLOBIN (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Kavanaugh, J.S, Arnone, A. | | 登録日 | 1997-12-08 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | High-resolution crystal structures of human hemoglobin with mutations at tryptophan 37beta: structural basis for a high-affinity T-state,.
Biochemistry, 37, 1998
|
|
1A0U
 
 | | HEMOGLOBIN (VAL BETA1 MET) MUTANT | | 分子名称: | HEMOGLOBIN (ALPHA CHAIN), HEMOGLOBIN (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Kavanaugh, J.S, Arnone, A. | | 登録日 | 1997-12-08 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | High-resolution crystal structures of human hemoglobin with mutations at tryptophan 37beta: structural basis for a high-affinity T-state,.
Biochemistry, 37, 1998
|
|
1A0Z
 
 | | HEMOGLOBIN (VAL BETA1 MET) MUTANT | | 分子名称: | HEMOGLOBIN (ALPHA CHAIN), HEMOGLOBIN (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Kavanaugh, J.S, Arnone, A. | | 登録日 | 1997-12-08 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | High-resolution crystal structures of human hemoglobin with mutations at tryptophan 37beta: structural basis for a high-affinity T-state,.
Biochemistry, 37, 1998
|
|
1A00
 
 | | HEMOGLOBIN (VAL BETA1 MET, TRP BETA37 TYR) MUTANT | | 分子名称: | HEMOGLOBIN (ALPHA CHAIN), HEMOGLOBIN (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Kavanaugh, J.S, Arnone, A. | | 登録日 | 1997-12-08 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | High-resolution crystal structures of human hemoglobin with mutations at tryptophan 37beta: structural basis for a high-affinity T-state,.
Biochemistry, 37, 1998
|
|
2CNP
 
 | |
2HGH
 
 | |
1DSI
 
 | | Solution structure of a duocarmycin sa-indole-alkylated dna dupleX | | 分子名称: | 4-HYDROXY-6-(1H-INDOLE-2-CARBONYL)-8-METHYL-3,6,7,8-TETRAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, DNA (5'-D(*GP*AP*CP*TP*AP*AP*TP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*AP*TP*TP*AP*GP*TP*C)-3') | | 著者 | Schnell, J.R, Ketchem, R.R, Boger, D.L, Chazin, W.J. | | 登録日 | 1998-07-29 | | 公開日 | 1998-08-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Binding-Induced Activation of DNA Alkylation by Duocarmycin SA: Insights from the Structure of an Indole Derivative-DNA Adduct
J.Am.Chem.Soc., 121, 1999
|
|
