3I4L
 
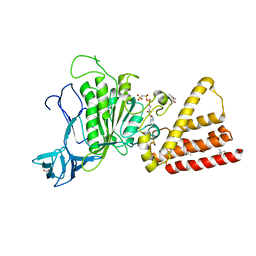 | | Structural characterization for the nucleotide binding ability of subunit A with AMP-PNP of the A1AO ATP synthase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, A-TYPE ATP SYNTHASE CATALYTIC SUBUNIT A, ... | | 著者 | Manimekalai, S.M.S, Kumar, A, Balakrishna, A.M, Jeyakanthan, J, Gruber, G. | | 登録日 | 2009-07-01 | | 公開日 | 2010-01-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Nucleotide binding states of subunit A of the A-ATP synthase and the implication of P-loop switch in evolution.
J.Mol.Biol., 396, 2010
|
|
8I47
 
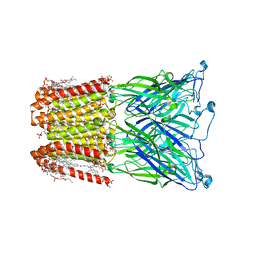 | |
8I41
 
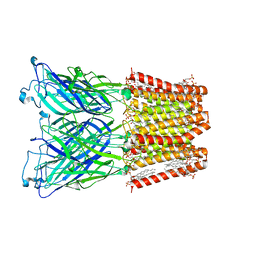 | |
8I42
 
 | |
8I48
 
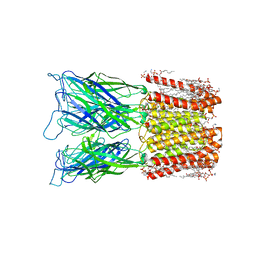 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in closed state | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | 著者 | Bharambe, N, Li, Z, Basak, S. | | 登録日 | 2023-01-18 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
8JJ3
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 2.5 | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | 著者 | Bharambe, N, Li, Z, Basak, S. | | 登録日 | 2023-05-29 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (2.6476 Å) | | 主引用文献 | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
8WCR
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in open state | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | 著者 | Bharambe, N, Li, Z, Basak, S. | | 登録日 | 2023-09-13 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
8WCQ
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in intermediate state | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | 著者 | Bharambe, N, Li, Z, Basak, S. | | 登録日 | 2023-09-13 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
2KK7
 
 | |
5B8A
 
 | | Crystal structure of oxidized chimeric EcAhpC1-186-YFSKHN | | 分子名称: | Alkyl hydroperoxide reductase subunit C,Peroxiredoxin-2, GLYCEROL, SULFATE ION | | 著者 | Kamariah, N, Sek, M.F, Eisenhaber, B, Eisenhaber, F, Gruber, G. | | 登録日 | 2016-06-14 | | 公開日 | 2017-02-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Transition steps in peroxide reduction and a molecular switch for peroxide robustness of prokaryotic peroxiredoxins.
Sci Rep, 6, 2016
|
|
5B8B
 
 | | Crystal structure of reduced chimeric E.coli AhpC1-186-YFSKHN | | 分子名称: | Alkyl hydroperoxide reductase subunit C,Peroxiredoxin-2, SULFATE ION | | 著者 | Kamariah, N, Sek, M.F, Eisenhaber, B, Eisenhaber, F, Gruber, G. | | 登録日 | 2016-06-14 | | 公開日 | 2017-02-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Transition steps in peroxide reduction and a molecular switch for peroxide robustness of prokaryotic peroxiredoxins.
Sci Rep, 6, 2016
|
|
3SSA
 
 | | Crystal structure of subunit B mutant N157T of the A1AO ATP synthase | | 分子名称: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Sundararaman, L, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | 登録日 | 2011-07-08 | | 公開日 | 2012-09-05 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of subunit A mutants H156A, N157A and N157T of the A1AO ATP synthase
To be Published
|
|
3TGW
 
 | | Crystal structure of subunit B mutant H156A of the A1AO ATP synthase | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, ... | | 著者 | Tadwal, V.S, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | 登録日 | 2011-08-18 | | 公開日 | 2012-09-05 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure of subunit A mutants H156A, N157A and N157T of the A1AO ATP synthase
To be Published
|
|
3TIV
 
 | | Crystal structure of subunit B mutant N157A of the A1AO ATP synthase | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CHLORIDE ION, ... | | 著者 | Tadwal, V.S, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | 登録日 | 2011-08-22 | | 公開日 | 2012-09-05 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure of subunit A mutants H156A, N157A and N157T of the A1AO ATP synthase
To be Published
|
|
4ITZ
 
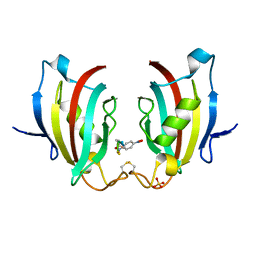 | |
3P20
 
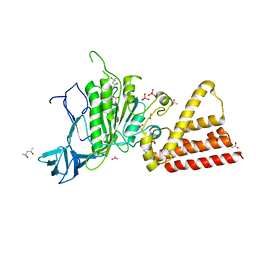 | | Crystal structure of vanadate bound subunit A of the A1AO ATP synthase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | 著者 | Manimekalai, M.S.S, Kumar, A, Jeyakanthan, J, Gruber, G. | | 登録日 | 2010-10-01 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | The transition-like state and Pi entrance into the catalytic a subunit of the biological engine A-ATP synthase.
J.Mol.Biol., 408, 2011
|
|
3QIA
 
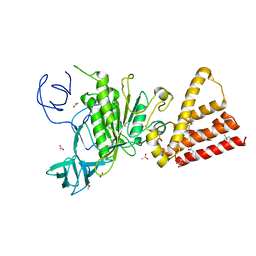 | | Crystal structure of P-loop G237A mutant of subunit A of the A1AO ATP synthase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | 著者 | Ragunathan, P, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | 登録日 | 2011-01-26 | | 公開日 | 2011-10-05 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
3QG1
 
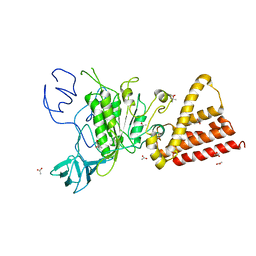 | | Crystal structure of P-loop G239A mutant of subunit A of the A1AO ATP synthase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, V-type ATP synthase alpha chain | | 著者 | Ragunathan, P, Manimekalai, M.S.S, Kumar, A, Jeyakanthan, J, Gruber, G. | | 登録日 | 2011-01-24 | | 公開日 | 2011-10-05 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
3QJY
 
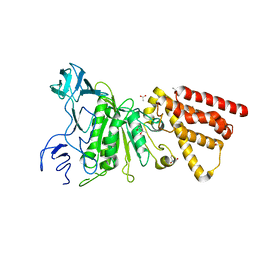 | | Crystal structure of P-loop G234A mutant of subunit A of the A1AO ATP synthase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | 著者 | Ragunathan, P, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | 登録日 | 2011-01-31 | | 公開日 | 2011-10-05 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
3NI6
 
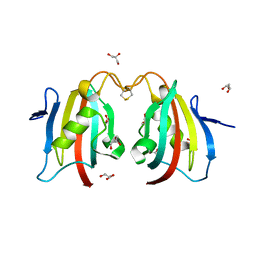 | |
4O5R
 
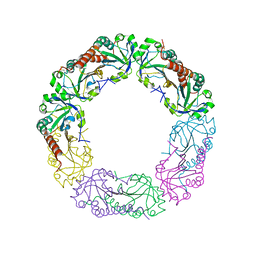 | | Crystal structure of Alkylhydroperoxide Reductase subunit C from E. coli | | 分子名称: | AhpC component, subunit of alkylhydroperoxide reductase | | 著者 | Dip, P.V, Kamariah, N, Manimekalai, M.S.S, Gruber, G, Eisenhaber, F, Eisenhaber, B. | | 登録日 | 2013-12-20 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.33 Å) | | 主引用文献 | Structure, mechanism and ensemble formation of the alkylhydroperoxide reductase subunits AhpC and AhpF from Escherichia coli
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4O5U
 
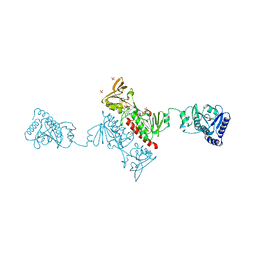 | | Crystal structure of Alkylhydroperoxide Reductase subunit F from E. coli at 2.65 Ang resolution | | 分子名称: | Alkyl hydroperoxide reductase subunit F, CADMIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Kamariah, N, Dip, P.V, Manimekalai, M.S.S, Gruber, G, Eisenhaber, F, Eisenhaber, B. | | 登録日 | 2013-12-20 | | 公開日 | 2014-11-05 | | 最終更新日 | 2015-02-25 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structure, mechanism and ensemble formation of the alkylhydroperoxide reductase subunits AhpC and AhpF from Escherichia coli
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4YKG
 
 | | Crystal Structure of the Alkylhydroperoxide Reductase subunit F (AhpF) with NAD+ from Escherichia coli | | 分子名称: | Alkyl hydroperoxide reductase subunit F, CADMIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kamariah, N, Manimekalai, M.S.S, Gruber, G, Eisenhaber, F, Eisenhaber, B. | | 登録日 | 2015-03-04 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystallographic and solution studies of NAD(+)- and NADH-bound alkylhydroperoxide reductase subunit F (AhpF) from Escherichia coli provide insight into sequential enzymatic steps
Biochim.Biophys.Acta, 1847, 2015
|
|
4YKF
 
 | | Crystal Structure of the Alkylhydroperoxide Reductase subunit F (AhpF) with NADH from Escherichia coli | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Alkyl hydroperoxide reductase subunit F, CADMIUM ION, ... | | 著者 | Kamariah, N, Manimekalai, M.S.S, Gruber, G, Eisenhaber, F, Eisenhaber, B. | | 登録日 | 2015-03-04 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystallographic and solution studies of NAD(+)- and NADH-bound alkylhydroperoxide reductase subunit F (AhpF) from Escherichia coli provide insight into sequential enzymatic steps
Biochim.Biophys.Acta, 1847, 2015
|
|
3DSR
 
 | | ADP in transition binding site in the subunit B of the energy converter A1Ao ATP synthase | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase beta chain | | 著者 | Kumar, A, Manimekalai, S.M.S, Balakrishna, A.M, Gruber, G. | | 登録日 | 2008-07-14 | | 公開日 | 2009-06-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of the nucleotide-binding subunit B of the energy producer A1A0 ATP synthase in complex with adenosine diphosphate
Acta Crystallogr.,Sect.D, 64, 2008
|
|
