5Z07
 
 | |
4HGO
 
 | |
4HGR
 
 | |
4HGN
 
 | |
5BSA
 
 | |
5BS7
 
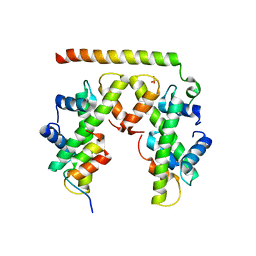 | | Structure of histone H3/H4 in complex with Spt2 | | 分子名称: | Histone H3.2, Histone H4, Protein SPT2 homolog, ... | | 著者 | Chen, S, Patel, D.J. | | 登録日 | 2015-06-01 | | 公開日 | 2015-07-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2.
Genes Dev., 29, 2015
|
|
7VJN
 
 | |
4HGP
 
 | |
4HGQ
 
 | |
4KSJ
 
 | |
4KSK
 
 | |
4KSL
 
 | |
3T7K
 
 | | Complex structure of Rtt107p and phosphorylated histone H2A | | 分子名称: | Histone H2A.1, Regulator of Ty1 transposition protein 107 | | 著者 | Li, X, Li, F, Wu, J, Shi, Y. | | 登録日 | 2011-07-30 | | 公開日 | 2012-02-15 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.028 Å) | | 主引用文献 | Structure of C-terminal Tandem BRCT Repeats of Rtt107 Protein Reveals Critical Role in Interaction with Phosphorylated Histone H2A during DNA Damage Repair
J.Biol.Chem., 287, 2012
|
|
3T7I
 
 | | Crystal structure of Se-Met Rtt107p (residues 820-1070) | | 分子名称: | Regulator of Ty1 transposition protein 107 | | 著者 | Li, X, Li, F, Wu, J, Shi, Y. | | 登録日 | 2011-07-30 | | 公開日 | 2012-02-15 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of C-terminal Tandem BRCT Repeats of Rtt107 Protein Reveals Critical Role in Interaction with Phosphorylated Histone H2A during DNA Damage Repair
J.Biol.Chem., 287, 2012
|
|
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | 分子名称: | 3C-like proteinase | | 著者 | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | 登録日 | 2020-12-24 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7VCQ
 
 | |
7VCL
 
 | |
7EO7
 
 | | Crystal structure of HCoV-NL63 3C-like protease in complex with an inhibitor Shikonin | | 分子名称: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | 著者 | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | 登録日 | 2021-04-21 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.24916625 Å) | | 主引用文献 | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7EO8
 
 | | Crystal structure of SARS coronavirus main protease in complex with an inhibitor Shikonin | | 分子名称: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | 著者 | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | 登録日 | 2021-04-21 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2808516 Å) | | 主引用文献 | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
3T7J
 
 | | Crystal structure of Rtt107p (residues 820-1070) | | 分子名称: | Regulator of Ty1 transposition protein 107 | | 著者 | Li, X, Li, F, Wu, J, Shi, Y. | | 登録日 | 2011-07-30 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.042 Å) | | 主引用文献 | Structure of C-terminal Tandem BRCT Repeats of Rtt107 Protein Reveals Critical Role in Interaction with Phosphorylated Histone H2A during DNA Damage Repair
J.Biol.Chem., 287, 2012
|
|
7X08
 
 | | S protein of SARS-CoV-2 in complex with 2G1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | 著者 | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | 登録日 | 2022-02-21 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Broad ultra-potent neutralization of SARS-CoV-2 variants by monoclonal antibodies specific to the tip of RBD.
Cell Discov, 8, 2022
|
|
6ZYT
 
 | | Monomeric streptavidin with a conjugated biotinylated pyrrolidine | | 分子名称: | 5-((3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)-N-((S)-pyrrolidin-3-yl)pentanamide, SULFATE ION, Streptavidin/Rhizavidin Hybrid | | 著者 | Nodling, A.R, Lipka-Lloyd, M, Tsai, Y.H, Rizkallah, P, Luk, L.Y.P, Jin, Y. | | 登録日 | 2020-08-03 | | 公開日 | 2021-08-18 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The role of streptavidin and its variants in catalysis by biotinylated secondary amines.
Org.Biomol.Chem., 19, 2021
|
|
8JF4
 
 | | The crystal structure of human AURKA kinase domain in complex with AURKA-compound 9 | | 分子名称: | 2-[4-[4-[bis(oxidanylidene)-$l^5-sulfanyl]oxyphenyl]carbonylpiperazin-1-yl]-6-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]-N-prop-2-ynyl-pyrimidine-4-carboxamide, Aurora kinase A | | 著者 | Zhu, C.J. | | 登録日 | 2023-05-17 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.89288354 Å) | | 主引用文献 | Global Reactivity Profiling of the Catalytic Lysine in Human Kinome for Covalent Inhibitor Development.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6A52
 
 | | Oxidase ChaP-H1 | | 分子名称: | FE (II) ION, dioxidase ChaP-H1 | | 著者 | Zhang, B, Ge, H.M. | | 登録日 | 2018-06-21 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
8JG8
 
 | |
