3OW7
 
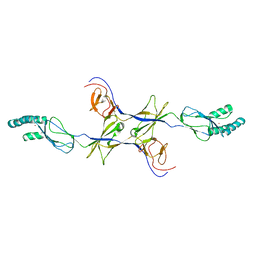 | |
3OPO
 
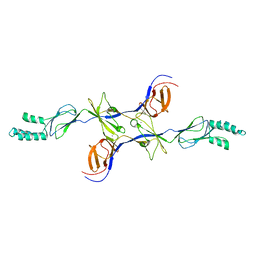 | |
3V53
 
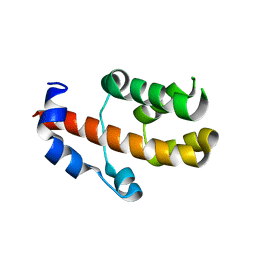 | | Crystal structure of human RBM25 | | 分子名称: | RNA-binding protein 25 | | 著者 | Gong, D.S. | | 登録日 | 2011-12-16 | | 公開日 | 2012-12-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure and functional characterization of the human RBM25 PWI domain and its flanking basic region
Biochem.J., 450, 2013
|
|
6LU5
 
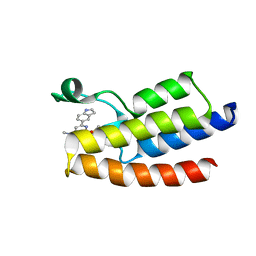 | | Crystal structure of BPTF-BRD with ligand DCBPin5 bound | | 分子名称: | 6-(1H-indol-5-yl)-N-methyl-2-methylsulfonyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | 著者 | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | 登録日 | 2020-01-25 | | 公開日 | 2021-04-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.86527729 Å) | | 主引用文献 | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
6LU6
 
 | | Crystal structure of BPTF-BRD with ligand DCBPin5-2 bound | | 分子名称: | 6-[1-[3-(dimethylamino)propyl]indol-5-yl]-2-methylsulfonyl-N-propyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | 著者 | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | 登録日 | 2020-01-26 | | 公開日 | 2021-04-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.970063 Å) | | 主引用文献 | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
1BVN
 
 | | PIG PANCREATIC ALPHA-AMYLASE IN COMPLEX WITH THE PROTEINACEOUS INHIBITOR TENDAMISTAT | | 分子名称: | CALCIUM ION, CHLORIDE ION, PROTEIN (ALPHA-AMYLASE), ... | | 著者 | Machius, M, Wiegand, G, Epp, O, Huber, R. | | 登録日 | 1998-09-16 | | 公開日 | 1998-09-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The crystal structure of porcine pancreatic alpha-amylase in complex with the microbial inhibitor Tendamistat.
J.Mol.Biol., 247, 1995
|
|
1DJ8
 
 | |
6OWK
 
 | |
1NDM
 
 | |
8TS6
 
 | |
3FLL
 
 | | Crystal structure of E55Q mutant of nitrophorin 4 | | 分子名称: | AMMONIA, Nitrophorin-4, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Montfort, W.R, Weichsel, A. | | 登録日 | 2008-12-18 | | 公開日 | 2009-02-10 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Effect of mutation of carboxyl side-chain amino acids near the heme on the midpoint potentials and ligand binding constants of nitrophorin 2 and its NO, histamine, and imidazole complexes.
J.Am.Chem.Soc., 131, 2009
|
|
1PIF
 
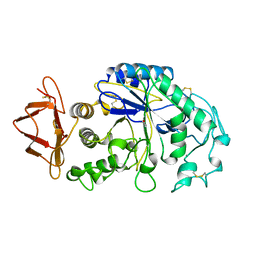 | | PIG ALPHA-AMYLASE | | 分子名称: | ALPHA-AMYLASE, CALCIUM ION, CHLORIDE ION | | 著者 | Machius, M, Vertesy, L, Huber, R, Wiegand, G. | | 登録日 | 1996-06-15 | | 公開日 | 1996-12-07 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Carbohydrate and protein-based inhibitors of porcine pancreatic alpha-amylase: structure analysis and comparison of their binding characteristics.
J.Mol.Biol., 260, 1996
|
|
3NVM
 
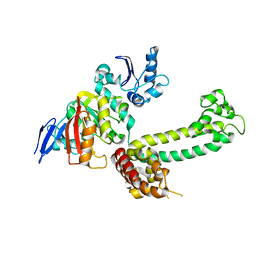 | |
1PIG
 
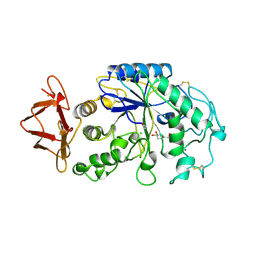 | | PIG PANCREATIC ALPHA-AMYLASE COMPLEXED WITH THE OLIGOSACCHARIDE V-1532 | | 分子名称: | 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, 5-HYDROXYMETHYL-CHONDURITOL, ... | | 著者 | Machius, M, Vertesy, L, Huber, R, Wiegand, G. | | 登録日 | 1996-06-15 | | 公開日 | 1996-12-07 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Carbohydrate and protein-based inhibitors of porcine pancreatic alpha-amylase: structure analysis and comparison of their binding characteristics.
J.Mol.Biol., 260, 1996
|
|
3NVI
 
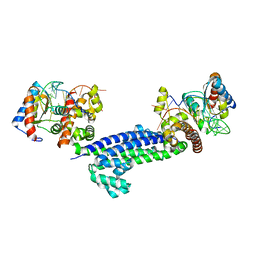 | |
5YS2
 
 | |
5YSL
 
 | | Crystal structure of antibody 1H1 Fab | | 分子名称: | 1H1 heavy chain, 1H1 light chain | | 著者 | Hu, X.L, Yang, F.L. | | 登録日 | 2017-11-14 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Two classes of protective antibodies against Pseudorabies virus variant glycoprotein B: Implications for vaccine design.
PLoS Pathog., 13, 2017
|
|
5JDK
 
 | |
3EE5
 
 | | Crystal structure of human M340H-Beta1,4-Galactosyltransferase-I (M340H-B4GAL-T1) in complex with GLCNAC-Beta1,3-Gal-Beta-Naphthalenemethanol | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, ... | | 著者 | Ramakrishnan, B, Qasba, P.K. | | 登録日 | 2008-09-04 | | 公開日 | 2009-01-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Deoxygenated Disaccharide Analogs as Specific Inhibitors of {beta}1-4-Galactosyltransferase 1 and Selectin-mediated Tumor Metastasis
J.Biol.Chem., 284, 2009
|
|
5WYH
 
 | |
3NMU
 
 | | Crystal Structure of substrate-bound halfmer box C/D RNP | | 分子名称: | 50S ribosomal protein L7Ae, Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, NOP5/NOP56 related protein, ... | | 著者 | Li, H, Xue, S, Wang, R. | | 登録日 | 2010-06-22 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.729 Å) | | 主引用文献 | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle.
Mol.Cell, 39, 2010
|
|
3NVK
 
 | | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle | | 分子名称: | 50S ribosomal protein L7Ae, Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, NOP5/NOP56 related protein, ... | | 著者 | Xue, S, Wang, R, Li, H. | | 登録日 | 2010-07-08 | | 公開日 | 2011-07-20 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.209 Å) | | 主引用文献 | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle.
Mol.Cell, 39, 2010
|
|
7C2J
 
 | | Crystal structure of nsp16-nsp10 heterodimer from SARS-CoV-2 in complex with SAM (without additional SAM during crystallization) | | 分子名称: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | 著者 | Lin, S, Chen, H, Ye, F, Chen, Z.M, Yang, F.L, Zheng, Y, Cao, Y, Qiao, J.X, Yang, S.Y, Lu, G.W. | | 登録日 | 2020-05-07 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.799 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Signal Transduct Target Ther, 5, 2020
|
|
7C2I
 
 | | Crystal structure of nsp16-nsp10 heterodimer from SARS-CoV-2 in complex with SAM (with additional SAM during crystallization) | | 分子名称: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | 著者 | Lin, S, Chen, H, Ye, F, Chen, Z.M, Yang, F.L, Zheng, Y, Cao, Y, Qiao, J.X, Yang, S.Y, Lu, G.W. | | 登録日 | 2020-05-07 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Signal Transduct Target Ther, 5, 2020
|
|
7JVM
 
 | |
