7EXZ
 
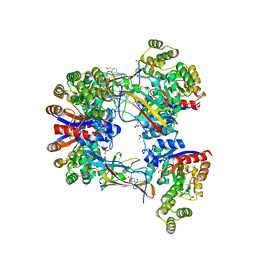 | | DgpB-DgpC complex apo 2.5 angstrom | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AP_endonuc_2 domain-containing protein, DgpB, ... | | 著者 | Mori, T, Senda, M, Senda, T, Abe, I. | | 登録日 | 2021-05-29 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7EXB
 
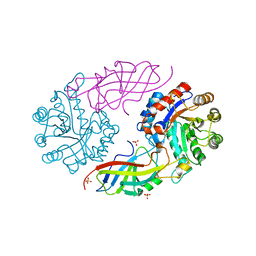 | | DfgA-DfgB complex apo 2.4 angstrom | | 分子名称: | DfgB, MANGANESE (II) ION, SULFATE ION, ... | | 著者 | Mori, T, Senda, M, Senda, T, Abe, I. | | 登録日 | 2021-05-26 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
6LTC
 
 | |
6LTD
 
 | |
6LTB
 
 | |
6LTA
 
 | |
7SQI
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and C14-crypto Acyl Carrier Protein, AcpP | | 分子名称: | Acyl carrier protein, Beta-ketoacyl-ACP synthase I, N-{2-[(2Z)-3-chlorotetradec-2-enamido]ethyl}-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | 著者 | Chen, A, Mindrebo, J.T, Davis, T.D, Noel, J.P, Burkart, M.D. | | 登録日 | 2021-11-05 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mechanism-based cross-linking probes capture the Escherichia coli ketosynthase FabB in conformationally distinct catalytic states.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7SZ9
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and C16:1-crypto Acyl Carrier Protein, AcpP | | 分子名称: | 3-oxoacyl-[acyl-carrier-protein] synthase 1, Acyl carrier protein, N~3~-{(2R)-4-[(dihydroxyphosphanyl)oxy]-2-hydroxy-3,3-dimethylbutanoyl}-N-(2-{[(9Z)-hexadec-9-enoyl]amino}ethyl)-beta-alaninamide, ... | | 著者 | Mindrebo, J.T, Chen, A, Noel, J.P, Burkart, M.D. | | 登録日 | 2021-11-26 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Mechanism-based cross-linking probes capture the Escherichia coli ketosynthase FabB in conformationally distinct catalytic states.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
3VNT
 
 | | Crystal Structure of the Kinase domain of Human VEGFR2 with a [1,3]thiazolo[5,4-b]pyridine derivative | | 分子名称: | 1,2-ETHANEDIOL, 2-chloro-3-(1-cyanocyclopropyl)-N-[5-({2-[(cyclopropylcarbonyl)amino][1,3]thiazolo[5,4-b]pyridin-5-yl}oxy)-2-fluorophenyl]benzamide, Vascular endothelial growth factor receptor 2 | | 著者 | Oki, H. | | 登録日 | 2012-01-17 | | 公開日 | 2012-04-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Design and synthesis of novel DFG-out RAF/vascular endothelial growth factor receptor 2 (VEGFR2) inhibitors. 1. Exploration of [5,6]-fused bicyclic scaffolds
J.Med.Chem., 55, 2012
|
|
5ZC9
 
 | | Crystal structure of the human eIF4A1-ATP analog-RocA-polypurine RNA complex | | 分子名称: | (1R,2R,3S,3aR,8bS)-6,8-dimethoxy-3a-(4-methoxyphenyl)-N,N-dimethyl-1,8b-bis(oxidanyl)-3-phenyl-2,3-dihydro-1H-cyclopenta[b][1]benzofuran-2-carboxamide, Eukaryotic initiation factor 4A-I, MAGNESIUM ION, ... | | 著者 | Iwasaki, W, Takahashi, M, Sakamoto, A, Iwasaki, S, Ito, T. | | 登録日 | 2018-02-16 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Translation Inhibitor Rocaglamide Targets a Bimolecular Cavity between eIF4A and Polypurine RNA.
Mol. Cell, 73, 2019
|
|
5ZT8
 
 | | SirB from Bacillus subtilis | | 分子名称: | Sirohydrochlorin ferrochelatase | | 著者 | Fujishiro, T. | | 登録日 | 2018-05-02 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of sirohydrochlorin ferrochelatase SirB: the last of the structures of the class II chelatase family.
Dalton Trans, 48, 2019
|
|
8GU3
 
 | |
7CUF
 
 | |
5ZT7
 
 | | SirB from Bacillus subtilis with Co2+ | | 分子名称: | COBALT (II) ION, Sirohydrochlorin ferrochelatase | | 著者 | Fujishiro, T. | | 登録日 | 2018-05-02 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Structure of sirohydrochlorin ferrochelatase SirB: the last of the structures of the class II chelatase family.
Dalton Trans, 48, 2019
|
|
1GA5
 
 | | CRYSTAL STRUCTURE OF THE ORPHAN NUCLEAR RECEPTOR REV-ERB(ALPHA) DNA-BINDING DOMAIN BOUND TO ITS COGNATE RESPONSE ELEMENT | | 分子名称: | 5'-D(*CP*AP*AP*CP*TP*AP*GP*GP*TP*CP*AP*CP*TP*AP*GP*GP*TP*CP*AP*G)-3', 5'-D(*CP*TP*GP*AP*CP*CP*TP*AP*GP*TP*GP*AP*CP*CP*TP*AP*GP*TP*(5IT)P*G)-3', ORPHAN NUCLEAR RECEPTOR NR1D1, ... | | 著者 | Sierk, M.L, Zhao, Q, Rastinejad, F. | | 登録日 | 2000-11-29 | | 公開日 | 2001-11-16 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | DNA Deformability as a Recognition Feature in the RevErb Response Element
Biochemistry, 40, 2001
|
|
7CUG
 
 | |
7C3G
 
 | | Crystal structure of human ALK2 kinase domain with R206H mutation in complex with a bicyclic pyrazole inhibitor RK-73134 | | 分子名称: | 1,2-ETHANEDIOL, Activin receptor type-1, SULFATE ION, ... | | 著者 | Sakai, N, Mishima-Tsumagari, C, Matsumoto, T, Shirouzu, M. | | 登録日 | 2020-05-12 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Novel bicyclic pyrazoles as potent ALK2 (R206H) inhibitors for the treatment of fibrodysplasia ossificans progressiva.
Bioorg.Med.Chem.Lett., 38, 2021
|
|
1H6I
 
 | |
7XZK
 
 | |
1GRL
 
 | | THE CRYSTAL STRUCTURE OF THE BACTERIAL CHAPERONIN GROEL AT 2.8 ANGSTROMS | | 分子名称: | GROEL (HSP60 CLASS) | | 著者 | Braig, K, Otwinowski, Z, Hegde, R, Boisvert, D.C, Joachimiak, A, Horwich, A.L, Sigler, P.B. | | 登録日 | 1995-03-07 | | 公開日 | 1995-10-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The crystal structure of the bacterial chaperonin GroEL at 2.8 A.
Nature, 371, 1994
|
|
1HLZ
 
 | | CRYSTAL STRUCTURE OF THE ORPHAN NUCLEAR RECEPTOR REV-ERB(ALPHA) DNA-BINDING DOMAIN BOUND TO ITS COGNATE RESPONSE ELEMENT | | 分子名称: | 5'-D(*CP*AP*AP*CP*TP*AP*GP*GP*TP*CP*AP*CP*TP*AP*GP*GP*TP*CP*AP*G)-3', 5'-D(*CP*TP*GP*AP*CP*CP*TP*AP*GP*TP*GP*AP*CP*CP*TP*AP*GP*TP*TP*G)-3', ORPHAN NUCLEAR RECEPTOR NR1D1, ... | | 著者 | Sierk, M.L, Zhao, Q, Rastinejad, F. | | 登録日 | 2000-12-04 | | 公開日 | 2001-11-16 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | DNA Deformability as a Recognition Feature in the RevErb Response Element
Biochemistry, 40, 2001
|
|
8HMM
 
 | | Crystal structure of AoRhaA | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bac_rhamnosid6H domain-containing protein | | 著者 | Makabe, K, Koseki, T. | | 登録日 | 2022-12-05 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Aspergillus oryzae alpha-l-rhamnosidase: Crystal structure and insight into the substrate specificity.
Proteins, 92, 2024
|
|
2PKP
 
 | | Crystal structure of 3-isopropylmalate dehydratase (leuD)from Methhanocaldococcus Jannaschii DSM2661 (MJ1271) | | 分子名称: | DI(HYDROXYETHYL)ETHER, Homoaconitase small subunit, ZINC ION | | 著者 | Jeyakanthan, J, Gayathri, D.R, Velmurugan, D, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-04-18 | | 公開日 | 2008-04-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Substrate specificity determinants of the methanogen homoaconitase enzyme: structure and function of the small subunit
Biochemistry, 49, 2010
|
|
7Y1G
 
 | | Crystal structure of human PRKACA complexed with DS01080522 | | 分子名称: | 1-chloranyl-~{N}-[(~{S})-(3-chloranyl-4-cyano-phenyl)-[(2~{R},4~{S})-4-oxidanylpyrrolidin-2-yl]methyl]-7-methoxy-isoquinoline-6-carboxamide, ZINC ION, cAMP-dependent protein kinase catalytic subunit alpha | | 著者 | Suzuki, M, Ubukata, O, Toyoda, A. | | 登録日 | 2022-06-08 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Novel protein kinase cAMP-Activated Catalytic Subunit Alpha (PRKACA) inhibitor shows anti-tumor activity in a fibrolamellar hepatocellular carcinoma model.
Biochem.Biophys.Res.Commun., 621, 2022
|
|
7VKF
 
 | | Reduced enzyme of FAD-dpendent Glucose Dehydrogenase complex with D-glucono-1,5-lactone at pH8.5 | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-glucono-1,5-lactone, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | 著者 | Nakajima, Y, Nishiya, Y, Ito, K. | | 登録日 | 2021-09-29 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Conformational change of catalytic residue in reduced enzyme of FAD-dependent Glucose Dehydrogenase at pH6.5
To Be Published
|
|
