8H0Y
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-112 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H12
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Locked-2 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.44681 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H15
 
 | | Structure of SARS-CoV-1 Spike Protein (S/native) at pH 5.5, Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.14182 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H0X
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-1 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.57 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
7VGC
 
 | |
2IF7
 
 | | Crystal Structure of NTB-A | | 分子名称: | CALCIUM ION, CHLORIDE ION, SLAM family member 6 | | 著者 | Cao, E, Ramagopal, U.A, Fedorov, A.A, Fedorov, E.V, Nathenson, S.G, Almo, S.C. | | 登録日 | 2006-09-20 | | 公開日 | 2006-10-17 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | NTB-A Receptor Crystal Structure: Insights into Homophilic Interactions in the Signaling Lymphocytic Activation Molecule Receptor Family.
Immunity, 25, 2006
|
|
7VGB
 
 | |
7FBJ
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing nanobody 17F6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, ... | | 著者 | Zhu, J, Xu, T, Feng, B, Liu, J. | | 登録日 | 2021-07-11 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
7FBK
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain N501Y mutant in complex with neutralizing nanobody 20G6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, Spike protein S1 | | 著者 | Zhu, J, Xu, T, Feng, B, Liu, J. | | 登録日 | 2021-07-11 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
8TXR
 
 | | E. coli ExoVII(H238A) | | 分子名称: | Exodeoxyribonuclease 7 large subunit, Exodeoxyribonuclease 7 small subunit | | 著者 | Liu, C, Berger, J.M. | | 登録日 | 2023-08-24 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of Escherichia coli exonuclease VII.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7XQP
 
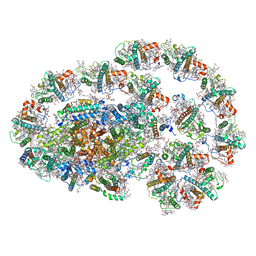 | | PSI-LHCI-LHCII-Lhcb9 supercomplex of Physcomitrella patens | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | 著者 | Zhang, S, Tang, K.L, Li, X.Y, Wang, W.D, Yan, Q.J, Shen, L.L, Kuang, T.Y, Han, G.Y, Shen, J.R, Zhang, X. | | 登録日 | 2022-05-08 | | 公開日 | 2023-04-26 | | 最終更新日 | 2023-06-07 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Structural insights into a unique PSI-LHCI-LHCII-Lhcb9 supercomplex from moss Physcomitrium patens.
Nat.Plants, 9, 2023
|
|
7YE9
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | 登録日 | 2022-07-05 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (4.17 Å) | | 主引用文献 | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YDY
 
 | | SARS-CoV-2 Spike (6P) in complex with 1 R1-32 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | 登録日 | 2022-07-04 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (4.75 Å) | | 主引用文献 | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YEG
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | 登録日 | 2022-07-05 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (3.73 Å) | | 主引用文献 | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YDI
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2, focused refinement of RBD region | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32, Light chain of R1-32, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | 登録日 | 2022-07-04 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (3.98 Å) | | 主引用文献 | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YE5
 
 | | SARS-CoV-2 Spike (6P) in complex with 2 R1-32 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | 登録日 | 2022-07-05 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (6.75 Å) | | 主引用文献 | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
4MPZ
 
 | |
9AUL
 
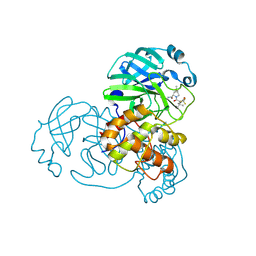 | | Structure of SARS-CoV-2 Mpro mutant (A173V,T304I)) in complex with Nirmatrelvir (PF-07321332) | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | 登録日 | 2024-02-29 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.421 Å) | | 主引用文献 | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUJ
 
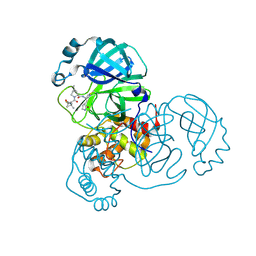 | | Structure of SARS-CoV-2 Mpro mutant (S144A) in complex with Nirmatrelvir (PF-07321332) | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | 登録日 | 2024-02-29 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.486 Å) | | 主引用文献 | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUM
 
 | | Structure of SARS-CoV-2 Mpro mutant (T21I,S144A,T304I) in complex with Nirmatrelvir (PF-07321332) | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | 登録日 | 2024-02-29 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.539 Å) | | 主引用文献 | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUK
 
 | | Structure of SARS-CoV-2 Mpro mutant (A173V) in complex with Nirmatrelvir (PF-07321332) | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | 登録日 | 2024-02-29 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUO
 
 | | Structure of SARS-CoV-2 Mpro mutant (L50F,T304I) | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | 登録日 | 2024-02-29 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.423 Å) | | 主引用文献 | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUN
 
 | | Structure of SARS-CoV-2 Mpro mutant (T21I,T304I) | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | 登録日 | 2024-02-29 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
7RFR
 
 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | 分子名称: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-(4-methoxy-1H-indole-2-carbonyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Gajiwala, K.S, Ferre, R.A, Liu, W, Stewart, A.E. | | 登録日 | 2021-07-14 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.626 Å) | | 主引用文献 | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
7RFU
 
 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | 分子名称: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-[N-(methanesulfonyl)-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | 登録日 | 2021-07-14 | | 公開日 | 2021-11-10 | | 最終更新日 | 2022-01-05 | | 実験手法 | X-RAY DIFFRACTION (2.498 Å) | | 主引用文献 | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
