8HVZ
 
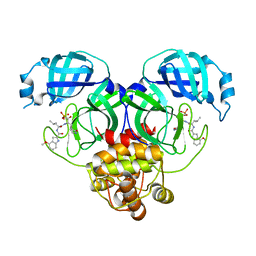 | |
8HVW
 
 | |
8IG4
 
 | |
8IG6
 
 | |
8IGA
 
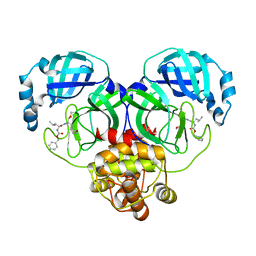 | |
8IG7
 
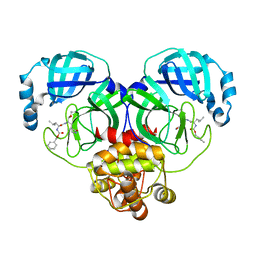 | |
8IG5
 
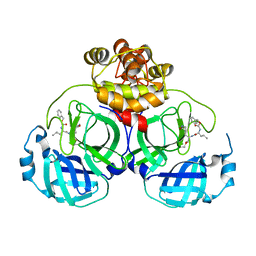 | | Crystal structure of SARS main protease in complex with GC376 | | 分子名称: | 3C-like proteinase nsp5, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | 著者 | Lin, C, Zhang, J, Li, J. | | 登録日 | 2023-02-20 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IG9
 
 | |
8IG8
 
 | |
8IGB
 
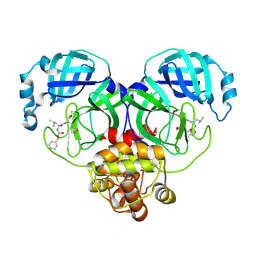 | |
4M02
 
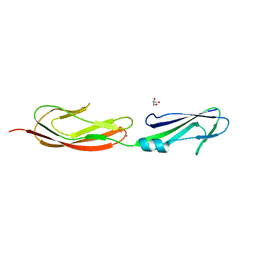 | | Middle fragment(residues 494-663) of the binding region of SraP | | 分子名称: | CALCIUM ION, GLYCEROL, Serine-rich adhesin for platelets | | 著者 | Yang, Y.H, Jiang, Y.L, Zhang, J, Wang, L, Chen, Y, Zhou, C.Z. | | 登録日 | 2013-08-01 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Structural Insights into SraP-Mediated Staphylococcus aureus Adhesion to Host Cells
Plos Pathog., 10, 2014
|
|
4M03
 
 | | C-terminal fragment(residues 576-751) of binding region of SraP | | 分子名称: | CALCIUM ION, Serine-rich adhesin for platelets | | 著者 | Yang, Y.H, Jiang, Y.L, Zhang, J, Wang, L, Chen, Y, Zhou, C.Z. | | 登録日 | 2013-08-01 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structural Insights into SraP-Mediated Staphylococcus aureus Adhesion to Host Cells
Plos Pathog., 10, 2014
|
|
4M00
 
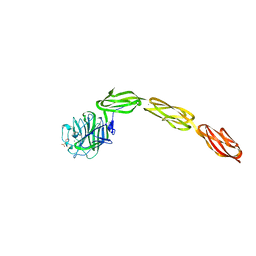 | | Crystal structure of the ligand binding region of staphylococcal adhesion SraP | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Serine-rich adhesin for platelets, ... | | 著者 | Yang, Y.H, Jiang, Y.L, Zhang, J, Wang, L, Chen, Y, Zhou, C.Z. | | 登録日 | 2013-08-01 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural Insights into SraP-Mediated Staphylococcus aureus Adhesion to Host Cells
Plos Pathog., 10, 2014
|
|
4M01
 
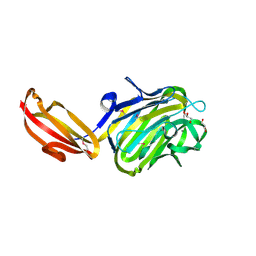 | | N terminal fragment(residues 245-575) of binding region of SraP | | 分子名称: | CALCIUM ION, GLYCEROL, Serine-rich adhesin for platelets | | 著者 | Yang, Y.H, Jiang, Y.L, Zhang, J, Wang, L, Chen, Y, Zhou, C.Z. | | 登録日 | 2013-08-01 | | 公開日 | 2014-06-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Insights into SraP-Mediated Staphylococcus aureus Adhesion to Host Cells
Plos Pathog., 10, 2014
|
|
4NTJ
 
 | | Structure of the human P2Y12 receptor in complex with an antithrombotic drug | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, P2Y purinoceptor 12,Soluble cytochrome b562,P2Y purinoceptor 12, ... | | 著者 | Zhang, K, Zhang, J, Gao, Z.-G, Zhang, D, Zhu, L, Han, G.W, Moss, S.M, Paoletta, S, Kiselev, E, Lu, W, Fenalti, G, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | 登録日 | 2013-12-02 | | 公開日 | 2014-03-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Structure of the human P2Y12 receptor in complex with an antithrombotic drug
Nature, 509, 2014
|
|
2AX5
 
 | | Solution Structure of Urm1 from Saccharomyces Cerevisiae | | 分子名称: | Hypothetical 11.0 kDa protein in FAA3-MAS3 intergenic region | | 著者 | Xu, J, Huang, H, Zhang, J, Wu, J, Shi, Y. | | 登録日 | 2005-09-03 | | 公開日 | 2006-06-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of Urm1 and its implications for the origin of protein modifiers.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5ITU
 
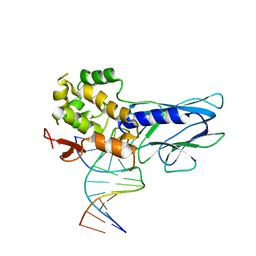 | | Crystal Structure of Human NEIL1(242K) bound to duplex DNA containing THF | | 分子名称: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*CP*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | 著者 | Zhu, C, Lu, L, Zhang, J, Yue, Z, Song, J, Zong, S, Liu, M, Stovicek, O, Gao, Y, Yi, C. | | 登録日 | 2016-03-17 | | 公開日 | 2016-07-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ITT
 
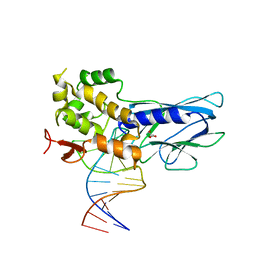 | | Crystal Structure of Human NEIL1 bound to duplex DNA containing THF | | 分子名称: | DNA (26-MER), Endonuclease 8-like 1, GLYCEROL | | 著者 | Zhu, C, Lu, L, Zhang, J, Yue, Z, Song, J, Zong, S, Liu, M, Stovicek, O, Gao, Y, Yi, C. | | 登録日 | 2016-03-17 | | 公開日 | 2016-07-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1YOP
 
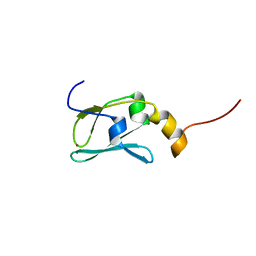 | | The solution structure of Kti11p | | 分子名称: | Kti11p, ZINC ION | | 著者 | Sun, J, Zhang, J, Wu, F, Xu, C, Li, S, Zhao, W, Wu, Z, Wu, J, Zhou, C.-Z, Shi, Y. | | 登録日 | 2005-01-28 | | 公開日 | 2005-04-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of Kti11p from Saccharomyces cerevisiae reveals a novel zinc-binding module.
Biochemistry, 44, 2005
|
|
5ITR
 
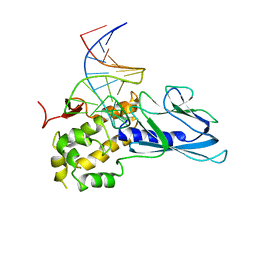 | | Crystal Structure of Human NEIL1(P2G) bound to duplex DNA containing THF | | 分子名称: | DNA (26-MER), Endonuclease 8-like 1 | | 著者 | Zhu, C, Lu, L, Zhang, J, Yue, Z, Song, J, Zong, S, Liu, M, Stovicek, O, Gao, Y, Yi, C. | | 登録日 | 2016-03-17 | | 公開日 | 2016-07-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ITQ
 
 | | Crystal Structure of Human NEIL1, Free Protein | | 分子名称: | Endonuclease 8-like 1 | | 著者 | Zhu, C, Lu, L, Zhang, J, Yue, Z, Song, J, Zong, S, Liu, M, Stovicek, O, Gao, Y, Yi, C. | | 登録日 | 2016-03-17 | | 公開日 | 2016-07-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ITY
 
 | | Crystal Structure of Human NEIL1(P2G) bound to duplex DNA containing Thymine Glycol | | 分子名称: | DNA (26-MER), Endonuclease 8-like 1, GLYCEROL | | 著者 | Zhu, C, Lu, L, Zhang, J, Yue, Z, Song, J, Zong, S, Liu, M, Stovicek, O, Gao, Y, Yi, C. | | 登録日 | 2016-03-17 | | 公開日 | 2016-07-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ITX
 
 | | Crystal Structure of Human NEIL1(P2G R242K) bound to duplex DNA containing Thymine Glycol | | 分子名称: | DNA (26-MER), Endonuclease 8-like 1 | | 著者 | Zhu, C, Lu, L, Zhang, J, Yue, Z, Song, J, Zong, S, Liu, M, Stovicek, O, Gao, Y, Yi, C. | | 登録日 | 2016-03-17 | | 公開日 | 2016-07-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8HUT
 
 | | Crystal structure of MERS main protease in complex with S217622 | | 分子名称: | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, ORF1a | | 著者 | Lin, C, Zhang, J, Li, J. | | 登録日 | 2022-12-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HUV
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with S217622 | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Zeng, P, Zhang, J, Li, J. | | 登録日 | 2022-12-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
