8PF2
 
 | |
4XCT
 
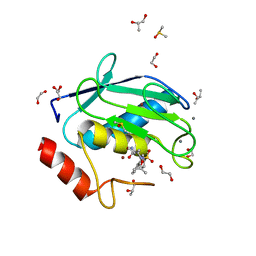 | | Crystal structure of a hydroxamate based inhibitor ARP101 (EN73) in complex with the MMP-9 catalytic domain. | | Descriptor: | (2S,3S)-butane-2,3-diol, (2~{R})-3-methyl-~{N}-oxidanylidene-2-[(4-phenylphenyl)sulfonyl-propan-2-yloxy-amino]butanamide, 1,2-ETHANEDIOL, ... | | Authors: | Stura, E.A, Tepshi, L, Nuti, E, Dive, V, Cassar-Lajeunesse, E, Vera, L, Rossello, A. | | Deposit date: | 2014-12-18 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | N-O-Isopropyl Sulfonamido-Based Hydroxamates as Matrix Metalloproteinase Inhibitors: Hit Selection and in Vivo Antiangiogenic Activity.
J.Med.Chem., 58, 2015
|
|
4WZV
 
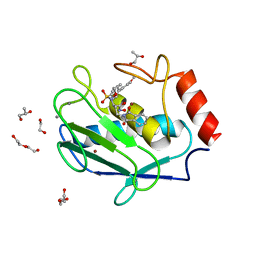 | | Crystal structure of a hydroxamate based inhibitor EN140 in complex with the MMP-9 catalytic domain | | Descriptor: | (2R)-4-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-N-hydroxy-2-{[(4'-methoxybiphenyl-4-yl)sulfonyl](propan-2-yloxy)amino}butanamide, 1,2-ETHANEDIOL, AZIDE ION, ... | | Authors: | Stura, E.A, Vera, L, Cassar-Lajeunesse, E, Nuti, E, Dive, V, Rossello, A. | | Deposit date: | 2014-11-20 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | N-O-Isopropyl Sulfonamido-Based Hydroxamates as Matrix Metalloproteinase Inhibitors: Hit Selection and in Vivo Antiangiogenic Activity.
J.Med.Chem., 58, 2015
|
|
8RND
 
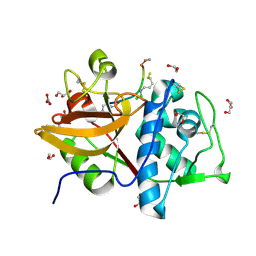 | | Cathepsin S in complex with NNPI-C10 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin S, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Petruzzella, A, Lau, K, Pojer, F, Oricchio, E. | | Deposit date: | 2024-01-09 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Antibody-peptide conjugates deliver covalent inhibitors blocking oncogenic cathepsins.
Nat.Chem.Biol., 20, 2024
|
|
8PI3
 
 | | Cathepsin S Y132D mutant in complex with NNPI-C10 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CADMIUM ION, ... | | Authors: | Petruzzella, A, Lau, K, Pojer, F, Oricchio, E. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Antibody-peptide conjugates deliver covalent inhibitors blocking oncogenic cathepsins.
Nat.Chem.Biol., 20, 2024
|
|
8OYD
 
 | |
4WBY
 
 | | tRNA-processing enzyme (apo form I) | | Descriptor: | GLYCEROL, Poly A polymerase, SULFATE ION | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2014-09-04 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Measurement of Acceptor-T Psi C Helix Length of tRNA for Terminal A76-Addition by A-Adding Enzyme.
Structure, 23, 2015
|
|
4WC5
 
 | | Structure of tRNA-processing enzyme complex 3 | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, Poly A polymerase, RNA (74-MER) | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2014-09-04 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Measurement of Acceptor-T Psi C Helix Length of tRNA for Terminal A76-Addition by A-Adding Enzyme.
Structure, 23, 2015
|
|
4WC4
 
 | | tRNA-processing enzyme complex 2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Poly A polymerase, RNA (74-MER) | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2014-09-04 | | Release date: | 2015-04-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Measurement of Acceptor-T Psi C Helix Length of tRNA for Terminal A76-Addition by A-Adding Enzyme.
Structure, 23, 2015
|
|
4WC6
 
 | | Structure of tRNA-processing enzyme complex 4 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Poly A polymerase, RNA (75-mer) | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2014-09-04 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Measurement of Acceptor-T Psi C Helix Length of tRNA for Terminal A76-Addition by A-Adding Enzyme.
Structure, 23, 2015
|
|
6N4D
 
 | | The crystal structure of neuramindase from A/canine/IL/11613/2015 (H3N2) influenza virus. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yang, H, Stevens, J. | | Deposit date: | 2018-11-19 | | Release date: | 2019-01-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Assessment of Molecular, Antigenic, and Pathological Features of Canine Influenza A(H3N2) Viruses That Emerged in the United States.
J. Infect. Dis., 216, 2017
|
|
6N4F
 
 | |
6HYC
 
 | |
6HPO
 
 | |
6J6Y
 
 | | FGFR4 D2 - Fab complex | | Descriptor: | Fab Heavy chain, Fab light chain, Fibroblast growth factor receptor 4 | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-08-07 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Preclinical Development of U3-1784, a Novel FGFR4 Antibody Against Cancer, and Avoidance of Its On-target Toxicity.
Mol.Cancer Ther., 18, 2019
|
|
6H0U
 
 | | Glycogen synthase kinase-3 beta (GSK3) complex with a covalent [1,2,4]triazolo[1,5-a][1,3,5]triazine inhibitor | | Descriptor: | (2~{R})-3-[7-azanyl-5-(cyclohexylamino)-[1,2,4]triazolo[1,5-a][1,3,5]triazin-2-yl]-2-cyano-propanamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Marcovich, I, Demitri, N, De Zorzi, R, Storici, P. | | Deposit date: | 2018-07-10 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Triazolotriazine-Based Dual GSK-3 beta /CK-1 delta Ligand as a Potential Neuroprotective Agent Presenting Two Different Mechanisms of Enzymatic Inhibition.
Chemmedchem, 14, 2019
|
|
3G3N
 
 | | PDE7A catalytic domain in complex with 3-(2,6-difluorophenyl)-2-(methylthio)quinazolin-4(3H)-one | | Descriptor: | 3-(2,6-difluorophenyl)-2-(methylthio)quinazolin-4(3H)-one, High affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A, MAGNESIUM ION, ... | | Authors: | Castano, T, Wang, H. | | Deposit date: | 2009-02-02 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis, structural analysis, and biological evaluation of thioxoquinazoline derivatives as phosphodiesterase 7 inhibitors
Chemmedchem, 4, 2009
|
|
4X0B
 
 | | Structure of tRNA-processing enzyme complex 7 | | Descriptor: | Poly A polymerase, RNA (77-MER) | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2014-11-21 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Measurement of Acceptor-T Psi C Helix Length of tRNA for Terminal A76-Addition by A-Adding Enzyme.
Structure, 23, 2015
|
|
4X0A
 
 | | Structure of tRNA-processing enzyme complex 6 | | Descriptor: | Poly A polymerase, RNA (73-MER) | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2014-11-21 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.505 Å) | | Cite: | Measurement of Acceptor-T Psi C Helix Length of tRNA for Terminal A76-Addition by A-Adding Enzyme.
Structure, 23, 2015
|
|
1PAH
 
 | |
6YTU
 
 | | Atomic-resolution structure of the coiled-coil dimerisation domain of human Arc | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Activity-regulated cytoskeleton-associated protein, CHLORIDE ION | | Authors: | Hallin, E.I, Touma, C, Bramham, C.R, Kursula, P. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Arc self-association and formation of virus-like capsids are mediated by an N-terminal helical coil motif.
Febs J., 288, 2021
|
|
6TQR
 
 | | The crystal structure of the MSP domain of human VAP-A in complex with the Phospho-FFAT motif of STARD3. | | Descriptor: | CHLORIDE ION, StAR-related lipid transfer protein 3, Vesicle-associated membrane protein-associated protein A | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
6TQT
 
 | | The crystal structure of the MSP domain of human MOSPD2. | | Descriptor: | 1,2-ETHANEDIOL, Motile sperm domain-containing protein 2, PHOSPHATE ION | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
5NMA
 
 | | Structure-activity relationship study of vitamin D analogs with oxolane group in their side chain | | Descriptor: | (1~{R},3~{S},5~{Z})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-7~{a}-methyl-1-[(1~{S})-1-[(2~{S},5~{S})-5-(2-oxidanylpropan-2-yl)oxolan-2-yl]ethyl]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N, Belorusova, A.Y. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-activity relationship study of vitamin D analogs with oxolane group in their side chain.
Eur J Med Chem, 134, 2017
|
|
6TQS
 
 | | The crystal structure of the MSP domain of human MOSPD2 in complex with the conventional FFAT motif of ORP1. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
