7K99
 
 | | Crystal Structure of P. aeruginosa LpxC with N-Hydroxyformamide inhibitor 19 | | Descriptor: | (hydroxy{(1S)-1-(methylsulfanyl)-2-[5-({4-[(morpholin-4-yl)methyl]phenyl}ethynyl)-1H-benzotriazol-1-yl]ethyl}amino)methanol, GLYCEROL, SULFATE ION, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2020-09-28 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | N-Hydroxyformamide LpxC inhibitors, their in vivo efficacy in a mouse Escherichia coli infection model, and their safety in a rat hemodynamic assay.
Bioorg.Med.Chem., 28, 2020
|
|
7YIM
 
 | | Cryo-EM structure of human Alpha-fetoprotein | | Descriptor: | Alpha-fetoprotein | | Authors: | Liu, N, Liu, K, Wu, C, Liu, Z, Li, M, Wang, J, Wang, H.W. | | Deposit date: | 2022-07-17 | | Release date: | 2023-01-18 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Uniform thin ice on ultraflat graphene for high-resolution cryo-EM.
Nat.Methods, 20, 2023
|
|
7K9A
 
 | | Crystal Structure of P. aeruginosa LpxC with N-Hydroxyformamide inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-hydroxy-N-[(1R)-2-{5-[(4-{[2-(hydroxymethyl)-1H-imidazol-1-yl]methyl}phenyl)ethynyl]-1H-benzotriazol-1-yl}-1-(methylsulfanyl)ethyl]formamide, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2020-09-29 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-Hydroxyformamide LpxC inhibitors, their in vivo efficacy in a mouse Escherichia coli infection model, and their safety in a rat hemodynamic assay.
Bioorg.Med.Chem., 28, 2020
|
|
7K88
 
 | | Crystal structure of bovine RPE65 in complex with hexaethylene glycol monooctyl ether | | Descriptor: | 3,6,9,12,15,18-hexaoxahexacosan-1-ol, FE (II) ION, Retinoid isomerohydrolase, ... | | Authors: | Kiser, P.D. | | Deposit date: | 2020-09-26 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational Alteration of Pharmacokinetics of Chiral Fluorinated and Deuterated Derivatives of Emixustat for Retinal Therapy.
J.Med.Chem., 64, 2021
|
|
7K8G
 
 | | Crystal structure of bovine RPE65 in complex with 4-fluoro-MB-004 and palmitate | | Descriptor: | (1R)-3-amino-1-{4-fluoro-3-[(2-propylpentyl)oxy]phenyl}propan-1-ol, FE (II) ION, PALMITIC ACID, ... | | Authors: | Kiser, P.D. | | Deposit date: | 2020-09-27 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational Alteration of Pharmacokinetics of Chiral Fluorinated and Deuterated Derivatives of Emixustat for Retinal Therapy.
J.Med.Chem., 64, 2021
|
|
7K89
 
 | |
7EBJ
 
 | | Apo structure of the mouse Trim66 PHD-Bromo dual domain | | Descriptor: | Tripartite motif-containing protein 66, ZINC ION | | Authors: | Wang, Z, Jiang, J. | | Deposit date: | 2021-03-09 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A TRIM66/DAX1/Dux axis suppresses the totipotent 2-cell-like state in murine embryonic stem cells.
Cell Stem Cell, 29, 2022
|
|
7DNK
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of 5G9 | | Descriptor: | Major capsid protein L1, The heavy chain of 5G9 Fab fragment, The light chain of 5G9 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (6.41 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
7DNL
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of A4B4 | | Descriptor: | Major capsid protein L1, The heavy chain of 2H3 Fab fragment, The light chain of A4B4 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
7D4B
 
 | | Crystal structure of 4-1BB in complex with a VHH | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wang, C. | | Deposit date: | 2020-09-23 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Generation of a safe and efficacious llama single-domain antibody fragment (vHH) targeting the membrane-proximal region of 4-1BB for engineering therapeutic bispecific antibodies for cancer.
J Immunother Cancer, 9, 2021
|
|
7CZD
 
 | | Crystal structure of PD-L1 in complex with a VHH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Programmed cell death 1 ligand 1, ... | | Authors: | Wang, C. | | Deposit date: | 2020-09-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Generation of a safe and efficacious llama single-domain antibody fragment (vHH) targeting the membrane-proximal region of 4-1BB for engineering therapeutic bispecific antibodies for cancer.
J Immunother Cancer, 9, 2021
|
|
7XGW
 
 | |
7DBF
 
 | |
7DC9
 
 | |
7DCA
 
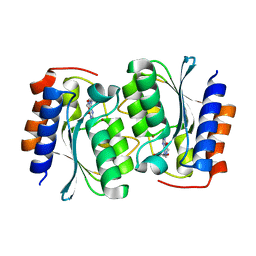 | | The structure of the Arabidopsis thaliana guanosine deaminase bound by xanthosine | | Descriptor: | 2,3-dihydroxanthosine, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-10-23 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The C-terminal loop of Arabidopsis thaliana guanosine deaminase is essential to catalysis.
Chem.Commun.(Camb.), 57, 2021
|
|
7DCB
 
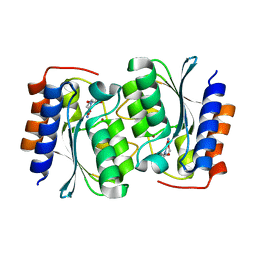 | |
7DCW
 
 | |
7DGW
 
 | | De novo designed protein H4A2S | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, de novo designed protein H4A2S | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DGU
 
 | | De novo designed protein H4A1R | | Descriptor: | de novo designed protein H4A1R | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DKK
 
 | | De novo design protein XM2H | | Descriptor: | De novo design protein XM2H | | Authors: | Bin, H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DKO
 
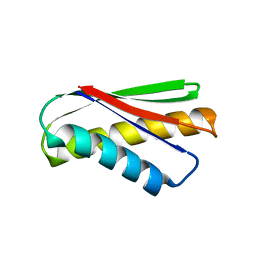 | | De novo design protein AM2M | | Descriptor: | de novo designed protein AM2M | | Authors: | Bin, H. | | Deposit date: | 2020-11-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DMF
 
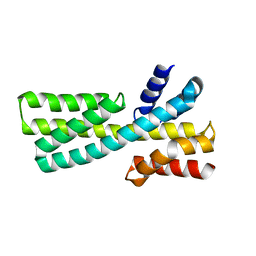 | |
7DGY
 
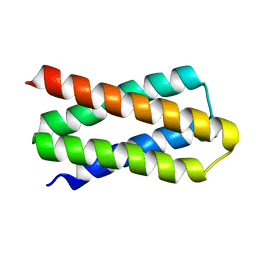 | | De novo designed protein H4C2R | | Descriptor: | de novo designed protein H4C2R | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DNH
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of 2H3 | | Descriptor: | Major capsid protein L1, The heavy chain of 2H3 Fab fragment, The light chain of 2H3 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
7EO0
 
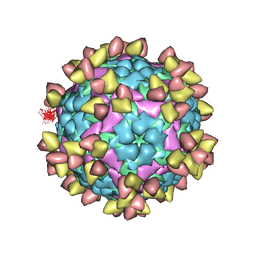 | | FOOT AND MOUTH DISEASE VIRUS O/TIBET/99-BOUND THE SINGLE CHAIN FRAGMEN ANTIBODY C4 | | Descriptor: | Ig heavy chain variable region, Ig lamda chain variable region, O/TIBET/99 VP1, ... | | Authors: | He, Y, Li, K. | | Deposit date: | 2021-04-21 | | Release date: | 2021-08-18 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Two Cross-Protective Antigen Sites on Foot-and-Mouth Disease Virus Serotype O Structurally Revealed by Broadly Neutralizing Antibodies from Cattle.
J.Virol., 95, 2021
|
|
