4N0G
 
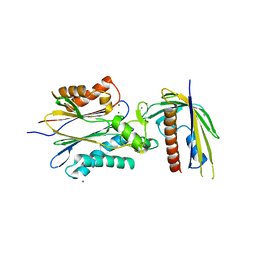 | | Crystal Structure of PYL13-PP2CA complex | | Descriptor: | Abscisic acid receptor PYL13, MAGNESIUM ION, Protein phosphatase 2C 37, ... | | Authors: | Li, W, Wang, L, Sheng, X, Yan, C, Zhou, R, Hang, J, Yin, P, Yan, N. | | Deposit date: | 2013-10-01 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Molecular basis for the selective and ABA-independent inhibition of PP2CA by PYL13
Cell Res., 23, 2013
|
|
7C9I
 
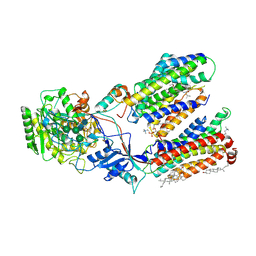 | | Human gamma-secretase in complex with small molecule L-685,458 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Guo, X, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-27 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of gamma-secretase inhibition and modulation by small molecule drugs.
Cell, 184, 2021
|
|
7C7Q
 
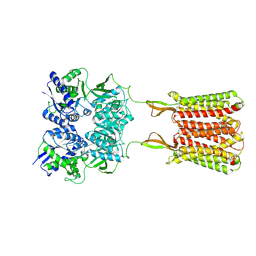 | | Cryo-EM structure of the baclofen/BHFF-bound human GABA(B) receptor in active state | | Descriptor: | (3S)-5,7-ditert-butyl-3-oxidanyl-3-(trifluoromethyl)-1-benzofuran-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Mao, C, Shen, C, Li, C, Shen, D, Xu, C, Zhang, S, Zhou, R, Shen, Q, Chen, L, Jiang, Z, Liu, J, Zhang, Y. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of inactive and active GABABreceptor.
Cell Res., 30, 2020
|
|
7C7S
 
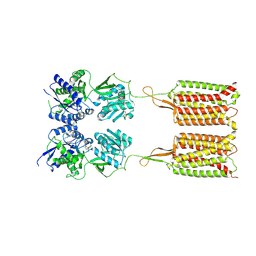 | | Cryo-EM structure of the CGP54626-bound human GABA(B) receptor in inactive state. | | Descriptor: | (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]amino}-2-hydroxypropyl]phosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Mao, C, Shen, C, Li, C, Shen, D, Xu, C, Zhang, S, Zhou, R, Shen, Q, Chen, L, Jiang, Z, Liu, J, Zhang, Y. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of inactive and active GABABreceptor.
Cell Res., 30, 2020
|
|
7D8X
 
 | | CryoEM structure of human gamma-secretase in complex with E2012 and L685458 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[(1S)-1-(4-fluorophenyl)ethyl]-3-[[3-methoxy-4-(4-methylimidazol-1-yl)phenyl]methylidene]piperidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Guo, X, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2020-10-11 | | Release date: | 2021-01-27 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of gamma-secretase inhibition and modulation by small molecule drugs.
Cell, 184, 2021
|
|
6LQG
 
 | | Human gamma-secretase in complex with small molecule Avagacestat | | Descriptor: | (2R)-2-[(4-chlorophenyl)sulfonyl-[[2-fluoranyl-4-(1,2,4-oxadiazol-3-yl)phenyl]methyl]amino]-5,5,5-tris(fluoranyl)pentanamide, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Guo, X, Lei, J, Shi, Y. | | Deposit date: | 2020-01-13 | | Release date: | 2021-01-27 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of gamma-secretase inhibition and modulation by small molecule drugs.
Cell, 184, 2021
|
|
6LR4
 
 | | Molecular basis for inhibition of human gamma-secretase by small molecule | | Descriptor: | (2S)-2-hydroxy-3-methyl-N-[(2S)-1-[[(5S)-3-methyl-4-oxo-2,5-dihydro-1H-3-benzazepin-5-yl]amino]-1-oxopropan-2-yl]butanamide, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Guo, X, Lei, J, Shi, Y. | | Deposit date: | 2020-01-15 | | Release date: | 2021-01-27 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of gamma-secretase inhibition and modulation by small molecule drugs.
Cell, 184, 2021
|
|
2EUA
 
 | | Structure and Mechanism of MenF, the Menaquinone-Specific Isochorismate Synthase from Escherichia Coli | | Descriptor: | D(-)-TARTARIC ACID, Menaquinone-specific isochorismate synthase | | Authors: | Kolappan, S, Kisker, C, Zwahlen, J, Zhou, R, Tonge, P.J. | | Deposit date: | 2005-10-28 | | Release date: | 2006-12-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lysine 190 is the catalytic base in MenF, the menaquinone-specific isochorismate synthase from Escherichia coli: implications for an enzyme family.
Biochemistry, 46, 2007
|
|
3NJE
 
 | | Structure of the Minor Pseudopilin XcpW from the Pseudomonas aeruginosa Type II Secretion System | | Descriptor: | General secretion pathway protein J | | Authors: | Franz, L.P, Dyer, D.H, Voulhoux, R, Forest, K.T. | | Deposit date: | 2010-06-17 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the minor pseudopilin XcpW from the Pseudomonas aeruginosa type II secretion system.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2I6Y
 
 | | Structure and Mechanism of Mycobacterium tuberculosis Salicylate Synthase, MbtI | | Descriptor: | Anthranilate synthase component I, putative | | Authors: | Zwahlen, J, Subramaniapillai, K, Zhou, R, Kisker, C, Tonge, P.J. | | Deposit date: | 2006-08-29 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of MbtI, the salicylate synthase from Mycobacterium tuberculosis
Biochemistry, 46, 2007
|
|
2KEP
 
 | | Solution structure of XcpT, the main component of the type 2 secretion system of Pseudomonas aeruginosa | | Descriptor: | General secretion pathway protein G | | Authors: | Alphonse, S, Durand, E, Douzi, B, Darbon, H, Filloux, A, Voulhoux, R, Bernard, C. | | Deposit date: | 2009-02-02 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Pseudomonas aeruginosa XcpT pseudopilin, a major component of the type II secretion system
J.Struct.Biol., 2009
|
|
7C22
 
 | |
2MWA
 
 | |
2MWF
 
 | |
2MWB
 
 | | FBP28 WW2 mutant W457F | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Macias, M.J, Scheraga, H, Sunol, D, Todorovski, T. | | Deposit date: | 2014-11-03 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding kinetics of WW domains with the united residue force field for bridging microscopic motions and experimental measurements.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2MWD
 
 | |
2MW9
 
 | |
2MWE
 
 | | NMR structure of FBP28 WW2 mutant Y438R, L453A DNDC | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Macias, M.J, Scheraga, H, Sunol, D, Todorovski, T. | | Deposit date: | 2014-11-04 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding kinetics of WW domains with the united residue force field for bridging microscopic motions and experimental measurements.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1CAP
 
 | | CONFORMATION AND MOLECULAR ORGANIZATION IN FIBERS OF THE CAPSULAR POLYSACCHARIDE FROM ESCHERICHIA COLI M41 MUTANT | | Descriptor: | alpha-D-mannopyranose-(1-3)-beta-D-glucopyranose-(1-3)-[4,6-O-[(1S)-1-carboxyethylidene]-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-4)]beta-D-glucopyranuronic acid-(1-3)-beta-D-galactopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-glucopyranose-(1-3)-[4,6-O-[(1S)-1-carboxyethylidene]-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-4)]beta-D-glucopyranuronic acid-(1-3)-beta-D-galactopyranose | | Authors: | Arnott, S. | | Deposit date: | 1978-05-23 | | Release date: | 1980-03-28 | | Last modified: | 2024-02-07 | | Method: | FIBER DIFFRACTION (3 Å) | | Cite: | Conformation and molecular organization in fibers of the capsular polysaccharide from Escherichia coli M41 mutant.
J.Mol.Biol., 109, 1977
|
|
7FBJ
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing nanobody 17F6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, ... | | Authors: | Zhu, J, Xu, T, Feng, B, Liu, J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
7FBK
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain N501Y mutant in complex with neutralizing nanobody 20G6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, Spike protein S1 | | Authors: | Zhu, J, Xu, T, Feng, B, Liu, J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
8IEM
 
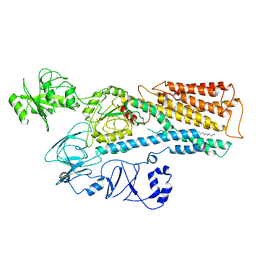 | | Cryo-EM structure of ATP13A2 in the E2P state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Polyamine-transporting ATPase 13A2, ... | | Authors: | Liu, Z.M, Mu, J.Q, Xue, C.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Conformational cycle of human polyamine transporter ATP13A2.
Nat Commun, 14, 2023
|
|
8IES
 
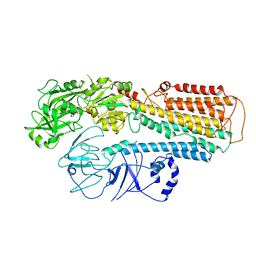 | | Cryo-EM structure of ATP13A2 in the E1P-ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Polyamine-transporting ATPase 13A2, ... | | Authors: | Liu, Z.M, Mu, J.Q, Xue, C.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Conformational cycle of human polyamine transporter ATP13A2.
Nat Commun, 14, 2023
|
|
8IEN
 
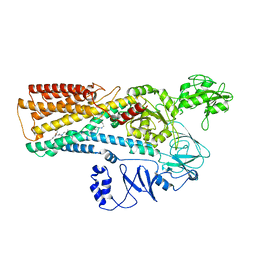 | | Cryo-EM structure of ATP13A2 in the E2-Pi state | | Descriptor: | MAGNESIUM ION, Polyamine-transporting ATPase 13A2, SPERMINE, ... | | Authors: | Liu, Z.M, Mu, J.Q, Xue, C.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Conformational cycle of human polyamine transporter ATP13A2.
Nat Commun, 14, 2023
|
|
8IER
 
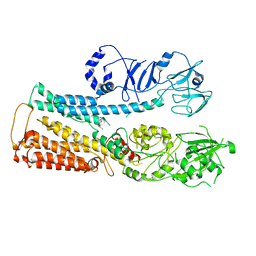 | |
