1W8X
 
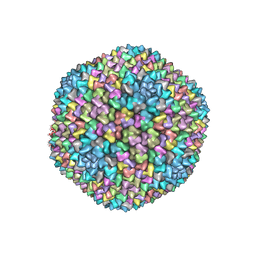 | | Structural analysis of PRD1 | | Descriptor: | MAJOR CAPSID PROTEIN (PROTEIN P3), PROTEIN P16, PROTEIN P30, ... | | Authors: | Abrescia, N.G.A, Cockburn, J.J.B, Grimes, J.M, Sutton, G.C, Diprose, J.M, Butcher, S.J, Fuller, S.D, San Martin, C, Burnett, R.M, Stuart, D.I, Bamford, D.H, Bamford, J.K.H. | | Deposit date: | 2004-10-01 | | Release date: | 2004-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Insights Into Assembly from Structural Analysis of Bacteriophage Prd1.
Nature, 432, 2004
|
|
1BYL
 
 | |
3MWS
 
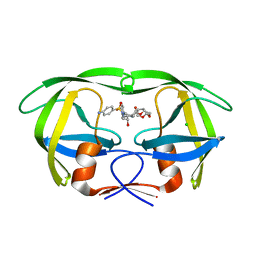 | | Crystal Structure of Group N HIV-1 Protease | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, HIV-1 Protease | | Authors: | Sayer, J.M, Agniswamy, J, Weber, I.T, Louis, J.M. | | Deposit date: | 2010-05-06 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Autocatalytic maturation, physical/chemical properties, and crystal structure of group N HIV-1 protease: relevance to drug resistance.
Protein Sci., 19, 2010
|
|
3RTP
 
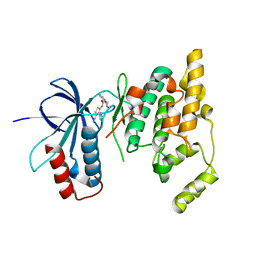 | | Design and synthesis of brain penetrant selective JNK inhibitors with improved pharmacokinetic properties for the prevention of neurodegeneration | | Descriptor: | Mitogen-activated protein kinase 10, N-[4-cyano-3-(1H-1,2,4-triazol-5-yl)thiophen-2-yl]-2-(2-oxo-3,4-dihydroquinolin-1(2H)-yl)acetamide | | Authors: | Bowers, S, Truong, A.P, Neitz, R.J, Hom, R.K, Sealy, J.M, Probst, G.D, Quincy, Q, Peterson, B, Chan, W, Galemmo Jr, R.A, Konradi, A.W, Sham, H.L, Pan, H, Lin, M, Yao, N, Artis, D.R, Zhang, H, Chen, L, Dryer, M, Samant, B, Zmolek, W, Wong, K, Lorentzen, C, Goldbach, E, Tonn, G, Quinn, K.P, Sauer, J, Wright, S, Powell, K, Ruslim, L, Ren, Z, Bard, F, Yednock, T.A, Griswold-Prenne, I. | | Deposit date: | 2011-05-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of brain penetrant selective JNK inhibitors with improved pharmacokinetic properties for the prevention of neurodegeneration.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1DAZ
 
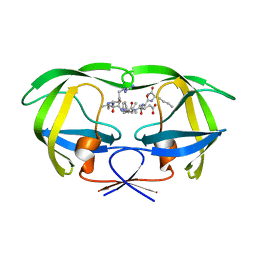 | | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease | | Descriptor: | HIV-1 PROTEASE (RETROPEPSIN), N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | Authors: | Mahalingam, B, Louis, J.M, Reed, C.C, Adomat, J.M, Krouse, J, Wang, Y.F, Harrison, R.W, Weber, I.T. | | Deposit date: | 1999-11-01 | | Release date: | 2000-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease.
Eur.J.Biochem., 263, 1999
|
|
1BAI
 
 | | Crystal structure of Rous sarcoma virus protease in complex with inhibitor | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE | | Authors: | Wu, J, Adomat, J.M, Ridky, T.W, Louis, J.M, Leis, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 1998-04-17 | | Release date: | 1999-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for specificity of retroviral proteases.
Biochemistry, 37, 1998
|
|
1DW6
 
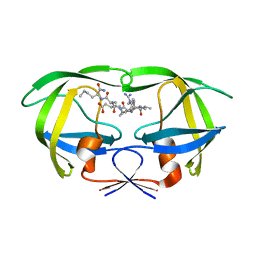 | | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease | | Descriptor: | HIV-1 PROTEASE, N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | Authors: | Mahalingam, B, Louis, J.M, Reed, C.C, Adomat, J.M, Krouse, J, Wang, Y.F, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-01-24 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease.
Eur.J.Biochem., 263, 1999
|
|
1ERI
 
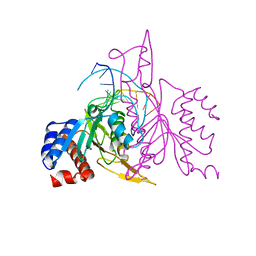 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE-DNA RECOGNITION COMPLEX: THE RECOGNITION NETWORK AND THE INTEGRATION OF RECOGNITION AND CLEAVAGE | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PROTEIN (ECO RI ENDONUCLEASE (E.C.3.1.21.4)) | | Authors: | Kim, Y, Grable, J.C, Love, R, Greene, P.J, Rosenberg, J.M. | | Deposit date: | 1994-05-18 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing.
Science, 249, 1990
|
|
3E4C
 
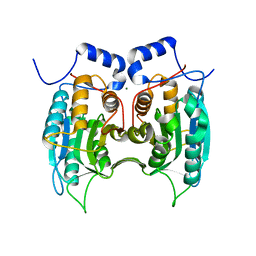 | | Procaspase-1 zymogen domain crystal structure | | Descriptor: | Caspase-1, MAGNESIUM ION | | Authors: | Elliott, J.M, Rouge, L, Wiesmann, C, Scheer, J.M. | | Deposit date: | 2008-08-11 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of procaspase-1 zymogen domain reveals insight into inflammatory caspase autoactivation
J.Biol.Chem., 284, 2009
|
|
6G4U
 
 | |
6G4I
 
 | |
6G4K
 
 | |
6G4V
 
 | | The solution NMR structure of [C18S,C24S]brevinin-1BYa in 33% trifluoroethanol | | Descriptor: | [C18S,C24S]brevinin-1BYa | | Authors: | Timmons, P.B, O'Flynn, D.P, Conlon, J.M, Hewage, C.M. | | Deposit date: | 2018-03-28 | | Release date: | 2019-10-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Insights into conformation and membrane interactions of the acyclic and dicarba-bridged brevinin-1BYa antimicrobial peptides.
Eur.Biophys.J., 48, 2019
|
|
6G4X
 
 | | The solution NMR structure of [C18S,C24S]brevinin-1BYa in sodium dodecyl sulphate micelles | | Descriptor: | [C18S,C24S]brevinin-1BYa | | Authors: | Timmons, P.B, O'Flynn, D.P, Conlon, J.M, Hewage, C.M. | | Deposit date: | 2018-03-28 | | Release date: | 2019-10-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Insights into conformation and membrane interactions of the acyclic and dicarba-bridged brevinin-1BYa antimicrobial peptides.
Eur.Biophys.J., 48, 2019
|
|
1PNC
 
 | |
3FK3
 
 | | Structure of the Yeats Domain, Yaf9 | | Descriptor: | Protein AF-9 homolog | | Authors: | Wang, A.Y, Schulze, J.M, Skordalakes, E, Berger, J.M, Rine, J, Kobor, M.S. | | Deposit date: | 2008-12-15 | | Release date: | 2009-10-27 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Asf1-like structure of the conserved Yaf9 YEATS domain and role in H2A.Z deposition and acetylation
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1PND
 
 | |
3G7C
 
 | | Structure of the Phosphorylation Mimetic of Occludin C-term Tail | | Descriptor: | Occludin | | Authors: | Tash, B.R, Sundstrom, J.M, Murakami, T, Flanagan, J.M, Bewley, M.C, Antonetii, D.A. | | Deposit date: | 2009-02-09 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and analysis of occludin phosphosites: a combined mass spectrometry and bioinformatics approach.
J.PROTEOME RES., 8, 2009
|
|
2CHY
 
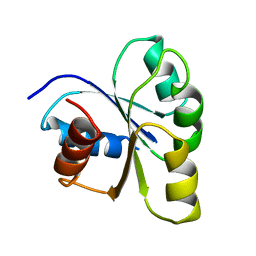 | | THREE-DIMENSIONAL STRUCTURE OF CHEY, THE RESPONSE REGULATOR OF BACTERIAL CHEMOTAXIS | | Descriptor: | CHEY | | Authors: | Mottonen, J.M, Stock, A.M, Stock, J.B, Schutt, C.E. | | Deposit date: | 1990-05-17 | | Release date: | 1990-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structure of CheY, the response regulator of bacterial chemotaxis.
Nature, 337, 1989
|
|
6I50
 
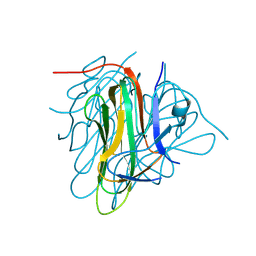 | |
2CPK
 
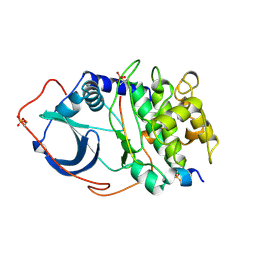 | | CRYSTAL STRUCTURE OF THE CATALYTIC SUBUNIT OF CYCLIC ADENOSINE MONOPHOSPHATE-DEPENDENT PROTEIN KINASE | | Descriptor: | PEPTIDE INHIBITOR 20-MER, cAMP-DEPENDENT PROTEIN KINASE, CATALYTIC SUBUNIT | | Authors: | Knighton, D.R, Zheng, J, Teneyck, L.F, Ashford, V.A, Xuong, N.-H, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 1992-10-21 | | Release date: | 1993-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase.
Science, 253, 1991
|
|
6I2I
 
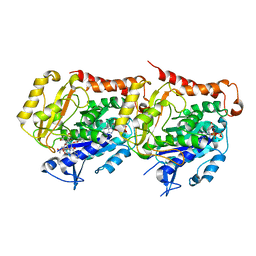 | | Refined 13pf Hela Cell Tubulin microtubule (EML4-NTD decorated) | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Atherton, J.M, Moores, C.A. | | Deposit date: | 2018-11-01 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mitotic phosphorylation by NEK6 and NEK7 reduces the microtubule affinity of EML4 to promote chromosome congression.
Sci.Signal., 12, 2019
|
|
2FDN
 
 | | 2[4FE-4S] FERREDOXIN FROM CLOSTRIDIUM ACIDI-URICI | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Dauter, Z, Wilson, K.S, Sieker, L.C, Meyer, J, Moulis, J.M. | | Deposit date: | 1997-10-01 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Atomic resolution (0.94 A) structure of Clostridium acidurici ferredoxin. Detailed geometry of [4Fe-4S] clusters in a protein.
Biochemistry, 36, 1997
|
|
1PYN
 
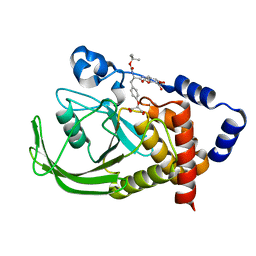 | | DUAL-SITE POTENT, SELECTIVE PROTEIN TYROSINE PHOSPHATASE 1B INHIBITOR USING A LINKED FRAGMENT STRATEGY AND A MALONATE HEAD ON THE FIRST SITE | | Descriptor: | 2-(4-{2-TERT-BUTOXYCARBONYLAMINO-2-[4-(3-HYDROXY-2-METHOXYCARBONYL-PHENOXY)-BUTYLCARBAMOYL]-ETHYL}-PHENOXY)-MALONIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Zhonghua, P, Lubben, T, Trevillyan, J.M, Stashko, M, Ballaron, S.J, Liang, H. | | Deposit date: | 2003-07-09 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and SAR of novel, potent and selective protein tyrosine phosphatase 1B inhibitors.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1POV
 
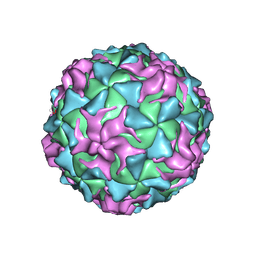 | |
