3V2H
 
 | | The crystal structure of D-beta-hydroxybutyrate dehydrogenase from Sinorhizobium meliloti | | Descriptor: | D-beta-hydroxybutyrate dehydrogenase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-12 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of D-beta-hydroxybutyrate dehydrogenase from Sinorhizobium meliloti
To be Published
|
|
7PZT
 
 | | Structure of the bacterial toxin, TecA, an asparagine deamidase from Alcaligenes faecalis. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Urea amidohydrolase | | Authors: | Dix, S.R, Aziz, A.A, Baker, P.J, Evans, C.A, Dickman, M.J, Farthing, R.J, King, Z.L.S, Nathan, S, Partridge, L.J, Raih, F.M, Sedelnikova, S.E, Thomas, M.S, Rice, D.W. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The structure of A. faecalis TecA provides insights into its role as an asparagine deamidase toxin which targets RhoA
To Be Published
|
|
5ABH
 
 | | Structure of GH84 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2R,3S,4R,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)-1-pentyl-pyrrolidin-2-yl]-N-methyl-ethanamide, CALCIUM ION, ... | | Authors: | Bergeron-Brlek, M, Goodwin-Tindall, J, Cekic, N, Varghese, V, Zandberg, W.F, Shan, X, Roth, C, Chan, S, Davies, G.J, Vocadlo, D.J, Britton, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable Oga Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5IZ7
 
 | | Cryo-EM structure of thermally stable Zika virus strain H/PF/2013 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, structural protein E, structural protein M | | Authors: | Kostyuchenko, V.A, Zhang, S, Fibriansah, G, Lok, S.M. | | Deposit date: | 2016-03-25 | | Release date: | 2016-05-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the thermally stable Zika virus
Nature, 533, 2016
|
|
3RYS
 
 | |
3S6J
 
 | | The crystal structure of a hydrolase from Pseudomonas syringae | | Descriptor: | CALCIUM ION, Hydrolase, haloacid dehalogenase-like family | | Authors: | Zhang, Z, Syed Ibrahim, B, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-05-25 | | Release date: | 2011-07-13 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | The crystal structure of a hydrolase from Pseudomonas syringae
To be Published
|
|
1G2X
 
 | | Sequence induced trimerization of krait PLA2: crystal structure of the trimeric form of krait PLA2 | | Descriptor: | PHOSPHOLIPASE A2 | | Authors: | Singh, G, Gourinath, S, Sharma, S, Bhanumathi, S, Paramsivam, M, Singh, T.P. | | Deposit date: | 2000-10-22 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sequence-induced trimerization of phospholipase A2: structure of a trimeric isoform of PLA2 from common krait (Bungarus caeruleus) at 2.5 A resolution.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
7VH2
 
 | |
7VGQ
 
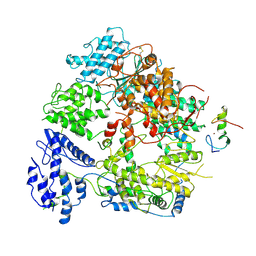 | | Cryo-EM structure of Machupo virus polymerase L in complex with matrix protein Z | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,RING finger protein Z, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Zhang, X, Ma, J, Zhang, S. | | Deposit date: | 2021-09-18 | | Release date: | 2021-09-29 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of Machupo virus polymerase in complex with matrix protein Z.
Nat Commun, 12, 2021
|
|
7VH1
 
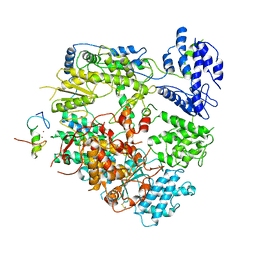 | | Cryo-EM structure of Machupo virus dimeric L-Z complex | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,RING finger protein Z, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Zhang, X, Ma, J, Zhang, S. | | Deposit date: | 2021-09-20 | | Release date: | 2021-09-29 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of Machupo virus polymerase in complex with matrix protein Z.
Nat Commun, 12, 2021
|
|
7VH3
 
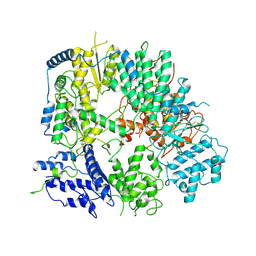 | |
3K6G
 
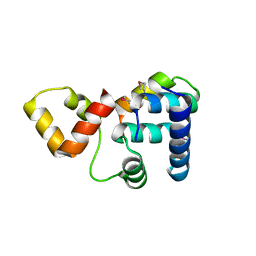 | | Crystal structure of Rap1 and TRF2 complex | | Descriptor: | Telomeric repeat-binding factor 2, Telomeric repeat-binding factor 2-interacting protein 1 | | Authors: | Chen, Y, Rai, R, Yang, Y.T, Zheng, H, Chang, S, Lei, M. | | Deposit date: | 2009-10-08 | | Release date: | 2010-10-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms.
Nat.Struct.Mol.Biol., 18, 2011
|
|
7D90
 
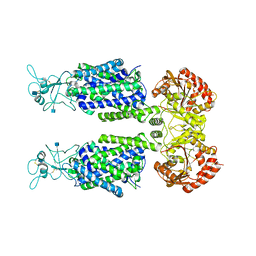 | | human potassium-chloride co-transporter KCC3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 3 | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-12 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
7D8Z
 
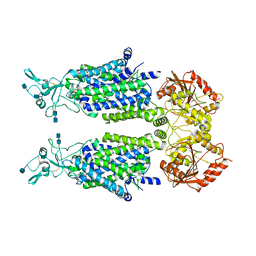 | | human potassium-chloride co-transporter KCC2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 2 | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-11 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
5ABE
 
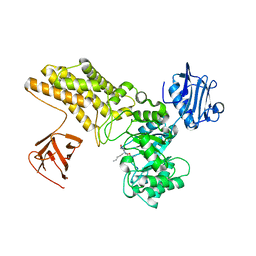 | | Structure of GH84 with ligand | | Descriptor: | 2-[(2S,3S,4R,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)-1-pentyl-pyrrolidin-2-yl]-N-methyl-ethanamide, CALCIUM ION, O-GLCNACASE BT_4395 | | Authors: | Bergeron-Brlek, M, Goodwin-Tindall, J, Cekic, N, Varghese, V, Zandberg, W.F, Shan, X, Roth, C, Chan, S, Davies, G.J, Vocadlo, D.J, Britton, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable Oga Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5IMR
 
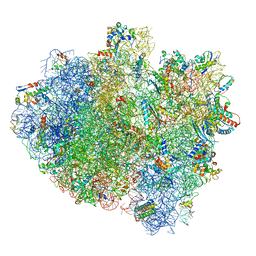 | | Structure of ribosome bound to cofactor at 5.7 angstrom resolution | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, V, Ero, R, Jian, G.K, Ahmed, T, Zhan, Y, Bhushan, S, Gao, Y.G. | | Deposit date: | 2016-03-06 | | Release date: | 2016-05-18 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structure of the GTP Form of Elongation Factor 4 (EF4) Bound to the Ribosome
J.Biol.Chem., 291, 2016
|
|
4RIS
 
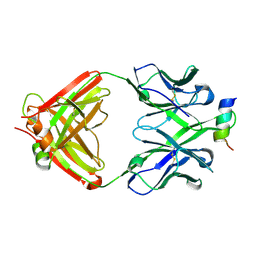 | | Structural Analysis of the Unmutated Ancestor of the HIV-1 Envelope V2 Region Antibody CH58 Isolated From an RV144 HIV-1 Vaccine Efficacy Trial Vaccinee and Associated with Decreased Transmission Risk | | Descriptor: | CH58-UA Fab heavy chain, CH58-UA Fab light chain, Envelope glycoprotein | | Authors: | Nicely, N.I, Wiehe, K, Kepler, T.B, Jaeger, F.H, Dennison, S.M, Liao, H.-X, Alam, S.M, Hwang, K.-K, Bonsignori, M, Rerks-Ngarm, S, Nitayaphan, S, Pitisuttithum, P, Kaewkungwal, J, Robb, M.L, O'Connell, R.J, Michael, N.L, Kim, J.H, Haynes, B.F. | | Deposit date: | 2014-10-07 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee.
EBioMedicine, 2, 2015
|
|
4RIR
 
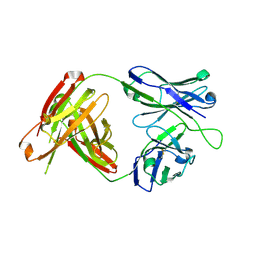 | | Structural Analysis of the Unmutated Ancestor of the HIV-1 Envelope V2 Region Antibody CH58 Isolated From an RV144 HIV-1 Vaccine Efficacy Trial Vaccinee and Associated with Decreased Transmission Risk | | Descriptor: | CH58-UA Fab heavy chain, CH58-UA Fab light chain | | Authors: | Nicely, N.I, Wiehe, K, Kepler, T.B, Jaeger, F.H, Dennison, S.M, Liao, H.-X, Alam, S.M, Hwang, K.-K, Bonsignori, M, Rerks-Ngarm, S, Nitayaphan, S, Pitisuttithum, P, Kaewkungwal, J, Robb, M.L, O'Connell, R.J, Michael, N.L, Kim, J.H, Haynes, B.F. | | Deposit date: | 2014-10-07 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee.
EBioMedicine, 2, 2015
|
|
1CQV
 
 | |
5IMQ
 
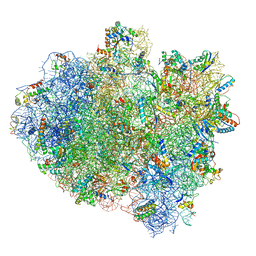 | | Structure of ribosome bound to cofactor at 3.8 angstrom resolution | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, V, Ero, R, Jian, G.K, Ahmed, T, Zhan, Y, Bhushan, S, Gao, Y.G. | | Deposit date: | 2016-03-06 | | Release date: | 2016-05-18 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the GTP Form of Elongation Factor 4 (EF4) Bound to the Ribosome
J.Biol.Chem., 291, 2016
|
|
7D99
 
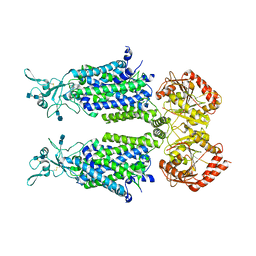 | | human potassium-chloride co-transporter KCC4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-12 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
3RAZ
 
 | |
1I4P
 
 | |
7VUL
 
 | |
1I4R
 
 | |
