5FMC
 
 | | Structure of D80A-fructofuranosidase from Xanthophyllomyces dendrorhous complexed with fructose and BIS-TRIS propane buffer | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ramirez-Escudero, M, Sanz-Aparicio, J. | | Deposit date: | 2015-11-02 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural Analysis of Beta-Fructofuranosidase from Xanthophyllomyces Dendrorhous Reveals Unique Features and the Crucial Role of N-Glycosylation in Oligomerization and Activity
J.Biol.Chem., 291, 2016
|
|
5FK7
 
 | |
5FK8
 
 | |
5FMB
 
 | |
8QCD
 
 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 GENE PRODUCT) IN COMPLEX WITH THE INHIBITOR 4,5,6,7-TETRABROMOBENZOTRIAZOLE | | Descriptor: | 1,2-ETHANEDIOL, 4,5,6,7-TETRABROMOBENZOTRIAZOLE, Casein kinase II subunit alpha' | | Authors: | Werner, C, Niefind, K. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Discovery and Exploration of Protein Kinase CK2 Binding Sites Using CK2alpha Cys336Ser as an Exquisite Crystallographic Tool
Kinases Phosphatases, 2023
|
|
8QBU
 
 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 GENE PRODUCT) IN COMPLEX WITH THE INHIBITOR CX-4945 AND THE ALPHA-D-POCKET LIGAND 3,4-DICHLORO PHENETHYLAMINE (DPA) | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,4-dichlorophenyl)ethanamine, 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, ... | | Authors: | Werner, C, Niefind, K. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Discovery and Exploration of Protein Kinase CK2 Binding Sites Using CK2alpha Cys336Ser as an Exquisite Crystallographic Tool
Kinases Phosphatases, 2023
|
|
3GF7
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum apoprotein | | Descriptor: | Glutaconyl-CoA decarboxylase subunit A, SULFATE ION | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-02-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
3GF3
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with glutaconyl-coA | | Descriptor: | CHLORIDE ION, CROTONYL COENZYME A, Glutaconyl-CoA decarboxylase subunit A | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-02-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
3GMA
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with glutaryl-CoA | | Descriptor: | Glutaconyl-CoA decarboxylase subunit A, glutaryl-coenzyme A | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-03-13 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
3GLM
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with crotonyl-coA | | Descriptor: | CHLORIDE ION, CROTONYL COENZYME A, Glutaconyl-CoA decarboxylase subunit A | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-03-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
6EMW
 
 | | Structure of S.aureus ClpC in complex with MecA | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit, ATP-dependent Clp protease ATP-binding subunit ClpC, Adapter protein MecA, ... | | Authors: | Carroni, M, Mogk, A, Bukau, B, Franke, K. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
6EM8
 
 | | S.aureus ClpC resting state, C2 symmetrised | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC | | Authors: | Carroni, M, Mogk, A. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
8OZQ
 
 | | In situ subtomogram average of Prototype Foamy Virus Env hexamer of trimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Calcraft, T, Nans, A, Rosenthal, P.B. | | Deposit date: | 2023-05-09 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Integrated cryoEM structure of a spumaretrovirus reveals cross-kingdom evolutionary relationships and the molecular basis for assembly and virus entry.
Cell, 187, 2024
|
|
8OZL
 
 | |
8OZN
 
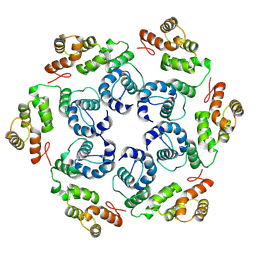 | |
8OZK
 
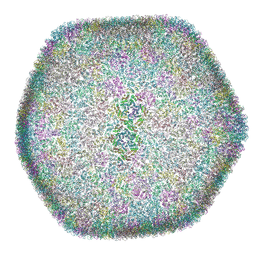 | |
8OZJ
 
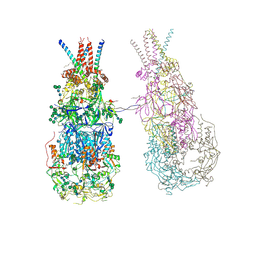 | | In situ cryoEM structure of Prototype Foamy Virus Env dimer of trimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Calcraft, T, Nans, A, Rosenthal, P.B. | | Deposit date: | 2023-05-09 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Integrated cryoEM structure of a spumaretrovirus reveals cross-kingdom evolutionary relationships and the molecular basis for assembly and virus entry.
Cell, 187, 2024
|
|
8OZP
 
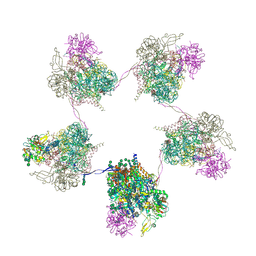 | | In situ subtomogram average of Prototype Foamy Virus Env pentamer of trimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Calcraft, T, Nans, A, Rosenthal, P.B. | | Deposit date: | 2023-05-09 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (11.9 Å) | | Cite: | Integrated cryoEM structure of a spumaretrovirus reveals cross-kingdom evolutionary relationships and the molecular basis for assembly and virus entry.
Cell, 187, 2024
|
|
8OZH
 
 | | In situ cryoEM structure of Prototype Foamy Virus Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Calcraft, T, Nans, A, Rosenthal, P.B. | | Deposit date: | 2023-05-09 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Integrated cryoEM structure of a spumaretrovirus reveals cross-kingdom evolutionary relationships and the molecular basis for assembly and virus entry.
Cell, 187, 2024
|
|
8OZM
 
 | |
6EM9
 
 | | S.aureus ClpC resting state, asymmetric map | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC | | Authors: | Carroni, M, Mogk, A, Bukau, B, Franke, K. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
4JNH
 
 | |
5M1H
 
 | |
5M1G
 
 | |
5MLU
 
 | | Crystal structure of the PFV GAG CBS bound to a mononucleosome | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B, ... | | Authors: | Pye, V.E, Maskell, D.P, Lesbats, P, Cherepanov, P. | | Deposit date: | 2016-12-07 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for spumavirus GAG tethering to chromatin.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
