6VIV
 
 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ000986192 | | Descriptor: | 2-[2-[(cyclohexylmethyl-$l^{3}-oxidanyl)carbonylamino]propan-2-yl]-~{N}-[2-(5-methoxy-4-oxidanyl-cyclohexa-1,3,5-trien-1-yl)ethyl]-5-oxidanyl-6-oxidanylidene-pyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-01-14 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
8QNI
 
 | |
8QNG
 
 | |
8QNH
 
 | | Crystal structure of the E3 ubiquitin ligase Cbl-b with an allosteric inhibitor (WO2020264398 Ex23) | | Descriptor: | 2-{3-[(1s,3R)-3-methyl-1-(4-methyl-4H-1,2,4-triazol-3-yl)cyclobutyl]phenyl}-6-{[(3S)-3-methylpiperidin-1-yl]methyl}-4-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-09-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Discovery of a Novel Benzodiazepine Series of Cbl-b Inhibitors for the Enhancement of Antitumor Immunity.
Acs Med.Chem.Lett., 14, 2023
|
|
6WLY
 
 | |
8QTK
 
 | | Crystal structure of CBL-b in complex with an allosteric inhibitor (compound 31) | | Descriptor: | 3-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-1-[(1S)-1-(1-methylpyrazol-4-yl)ethyl]-5-(trifluoromethyl)pyridin-2-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.873 Å) | | Cite: | Discovery, Optimization, and Biological Evaluation of Arylpyridones as Cbl-b Inhibitors.
J.Med.Chem., 67, 2024
|
|
6WLX
 
 | |
8QPD
 
 | | Structure of thioredoxin m from pea | | Descriptor: | Thioredoxin M-type, chloroplastic | | Authors: | Neira, J.L, Camara Artigas, A. | | Deposit date: | 2023-10-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure, dynamics and binding of thioredoxin m from Pisum sativum.
Int.J.Biol.Macromol., 262, 2024
|
|
6X0B
 
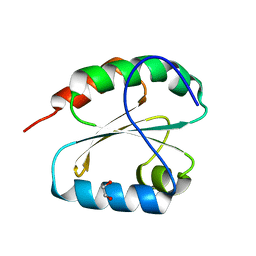 | |
6X2I
 
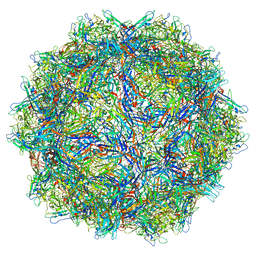 | |
8R74
 
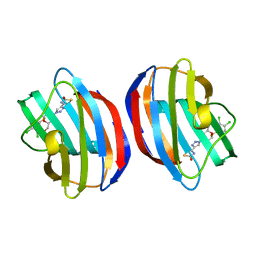 | | Galectin-1 in complex with thiogalactoside derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(3,4-dichlorophenyl)sulfanyl-6-(hydroxymethyl)-4-[4-(2-oxidanyl-1,3-thiazol-4-yl)-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-1 | | Authors: | Hakansson, M, Diehl, C, Peterson, K, Zetterberg, F, Nilsson, U. | | Deposit date: | 2023-11-23 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of the Selective and Orally Available Galectin-1 Inhibitor GB1908 as a Potential Treatment for Lung Cancer.
J.Med.Chem., 67, 2024
|
|
8QUS
 
 | |
8QU7
 
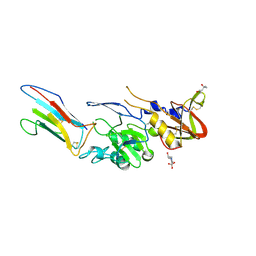 | |
8QIX
 
 | | Structure of Mycobacterium abscessus Phosphopantetheine adenylyltransferase in complex with inhibitor | | Descriptor: | 3-(3-methylindol-1-yl)-~{N}-(4-phenoxyphenyl)sulfonyl-propanamide, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, McCarthy, W.J, Coyne, A.G, Blundell, T.L. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | A Fragment-Based Competitive 19 F LB-NMR Platform For Hotspot-Directed Ligand Profiling.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8QID
 
 | | Structure of Mycobacterium abscessus Phosphopantetheine adenylyltransferase in complex with fragment | | Descriptor: | 1-phenyl-5-(trifluoromethyl)pyrazole-4-carboxylic acid, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, McCarthy, W.J, Coyne, A.G, Blundell, T.L. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A Fragment-Based Competitive 19 F LB-NMR Platform For Hotspot-Directed Ligand Profiling.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6X3N
 
 | | Co-structure of BTK kinase domain with L-005085737 inhibitor | | Descriptor: | 4-[8-azanyl-3-[(2~{S})-1-[4-(dimethylamino)butanoyl]pyrrolidin-2-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-(1,3-thiazol-2-yl)benzamide, 4-{8-amino-3-[(6R,8aS)-3-oxohexahydro-3H-[1,3]oxazolo[3,4-a]pyridin-6-yl]imidazo[1,5-a]pyrazin-1-yl}-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide, Tyrosine-protein kinase BTK | | Authors: | Fischmann, T.O. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
8QIY
 
 | | Structure of Mycobacterium abscessus Phosphopantetheine adenylyltransferase in complex with inhibitor | | Descriptor: | 1-(2-aminophenyl)-5-(trifluoromethyl)pyrazole-4-carboxylic acid, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, McCarthy, W.J, Coyne, A.G, Blundell, T.L. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.5149 Å) | | Cite: | A Fragment-Based Competitive 19 F LB-NMR Platform For Hotspot-Directed Ligand Profiling.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8QJ8
 
 | | Structure of Mycobacterium abscessus Phosphopantetheine adenylyltransferase in complex with inhibitor | | Descriptor: | 3-[3-(3-azanyl-2-cyano-phenyl)indol-1-yl]propanoic acid, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, McCarthy, W.J, Coyne, A.G, Blundell, T.L. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.538 Å) | | Cite: | A Fragment-Based Competitive 19 F LB-NMR Platform For Hotspot-Directed Ligand Profiling.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6WKK
 
 | | Phage G gp27 major capsid proteins and gp26 decoration proteins | | Descriptor: | Gp26 capsid decoration protein, Gp27 major capsid protein | | Authors: | Monroe, L, Gonzalez, B, Jiang, W, Kihara, D. | | Deposit date: | 2020-04-16 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Phage G Structure at 6.1 angstrom Resolution, Condensed DNA, and Host Identity Revision to a Lysinibacillus.
J.Mol.Biol., 432, 2020
|
|
6WLF
 
 | |
6WO1
 
 | |
8QPZ
 
 | | CryoEM structure of recombinant DeltaN7 alpha-synuclein in PBS | | Descriptor: | Alpha-synuclein | | Authors: | Thacker, D, Wilkinson, M, Dewison, K.M, Ranson, N.A, Brockwell, D.J, Radford, S.E. | | Deposit date: | 2023-10-03 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Residues 2 to 7 of alpha-synuclein regulate amyloid formation via lipid-dependent and lipid-independent pathways.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6X3O
 
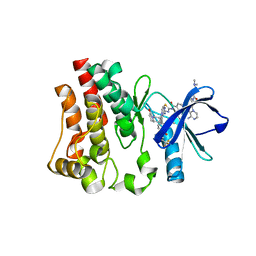 | | Co-structure of BTK kinase domain with L-005191930 inhibitor | | Descriptor: | 4-[8-azanyl-3-[(2~{S})-1-[4-(dimethylamino)butanoyl]pyrrolidin-2-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-(1,3-thiazol-2-yl)benzamide, 4-{8-amino-3-[(6R,8aS)-3-oxo-3,5,6,7,8,8a-hexahydroindolizin-6-yl]imidazo[1,5-a]pyrazin-1-yl}-3-methoxy-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide, Tyrosine-protein kinase BTK | | Authors: | Fischmann, T.O. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6X2K
 
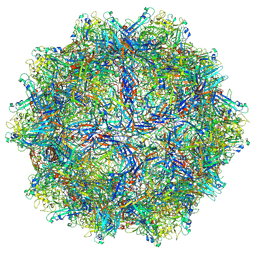 | |
6WJ5
 
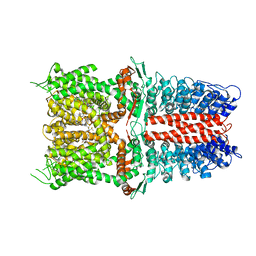 | | Structure of human TRPA1 in complex with inhibitor GDC-0334 | | Descriptor: | (4R,5S)-4-fluoro-1-[(4-fluorophenyl)sulfonyl]-5-methyl-N-({5-(trifluoromethyl)-2-[2-(trifluoromethyl)pyrimidin-5-yl]pyridin-4-yl}methyl)-L-prolinamide, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L, Arthur, C.P, Volgraf, M, Chen, H. | | Deposit date: | 2020-04-11 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A TRPA1 inhibitor suppresses neurogenic inflammation and airway contraction for asthma treatment.
J.Exp.Med., 218, 2021
|
|
