6PU7
 
 | | Human IDO1 in complex with compound 17 (N-{2-[(4-{N-[(7S)-4-fluorobicyclo[4.2.0]octa-1,3,5-trien-7-yl]-N'-hydroxycarbamimidoyl}-1,2,5-oxadiazol-3-yl)sulfanyl]ethyl}acetamide) | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-{2-[(4-{N-[(7S)-4-fluorobicyclo[4.2.0]octa-1,3,5-trien-7-yl]-N'-hydroxycarbamimidoyl}-1,2,5-oxadiazol-3-yl)sulfanyl]ethyl}acetamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-07-17 | | Release date: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of Amino-cyclobutarene-derived Indoleamine-2,3-dioxygenase 1 (IDO1) Inhibitors for Cancer Immunotherapy.
Acs Med.Chem.Lett., 10, 2019
|
|
5XW2
 
 | |
4RT4
 
 | | Crystal structure of Dpy30 complexed with Bre2 | | Descriptor: | Peptide from COMPASS component BRE2, Protein dpy-30 homolog | | Authors: | Zhang, H.M, Li, M, Chang, W.R. | | Deposit date: | 2014-11-12 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural implications of Dpy30 oligomerization for MLL/SET1 COMPASS H3K4 trimethylation
Protein Cell, 6, 2015
|
|
4RTA
 
 | |
5K25
 
 | |
5K23
 
 | |
5K24
 
 | |
8HYJ
 
 | |
5WYS
 
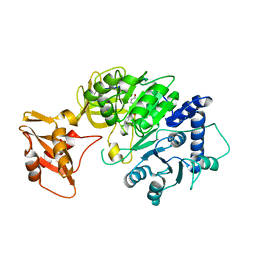 | | luciferase with inhibitor 3i | | Descriptor: | 5-[(3R)-3-(4-boranylphenyl)-3-oxidanyl-propyl]-2-oxidanyl-benzoic acid, Luciferin 4-monooxygenase | | Authors: | Gu, L, Su, J, Wang, F. | | Deposit date: | 2017-01-15 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Inhibiting Firefly Bioluminescence by Chalcones
Anal. Chem., 89, 2017
|
|
7PZJ
 
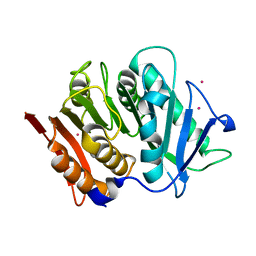 | | Structure of a bacteroidetal polyethylene terephthalate (PET) esterase | | Descriptor: | Lipase, POTASSIUM ION | | Authors: | Zang, H, Dierkes, R, Perez-Garcia, P, Weigert, S, Sternagel, S, Hallam, S.J, Applegate, V, Schumacher, J, Schott, T, Pleiss, J, Almeida, A, Hoecker, B, Smits, S.H, Schmitz, R.A, Chow, J, Streit, W.R. | | Deposit date: | 2021-10-12 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Bacteroidetes Aequorivita sp. and Kaistella jeonii Produce Promiscuous Esterases With PET-Hydrolyzing Activity.
Front Microbiol, 12, 2021
|
|
1RCS
 
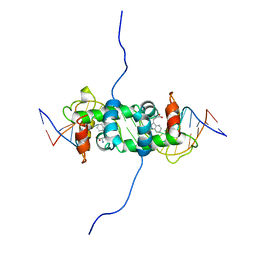 | | NMR STUDY OF TRP REPRESSOR-OPERATOR DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*TP*AP*GP*TP*AP*CP*G)-3'), TRP REPRESSOR, TRYPTOPHAN | | Authors: | Zhao, D, Zheng, Z. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the trp repressor-operator DNA complex.
J.Mol.Biol., 238, 1994
|
|
5YAT
 
 | |
4B5D
 
 | | Capitella teleta AChBP in complex with psychonicline (3-((2(S)- Azetidinyl)methoxy)-5-((1S,2R)-2-(2-hydroxyethyl)cyclopropyl)pyridine) | | Descriptor: | 2-[(1R,2S)-2-[5-[[(2S)-azetidin-2-yl]methoxy]pyridin-3-yl]cyclopropyl]ethanol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nys, M, Ulens, C. | | Deposit date: | 2012-08-03 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Insights Into the Structural Determinants Required for High- Affinity Binding of Chiral Cyclopropane-Containing Ligands to Alpha4Beta2-Nicotinic Acetylcholine Receptors: An Integrated Approach to Behaviorally Active Nicotinic Ligands.
J.Med.Chem., 55, 2012
|
|
8X5D
 
 | |
8QBJ
 
 | | Structure of mBaoJin at pH 4.6 | | Descriptor: | CHLORIDE ION, mBaoJin | | Authors: | Samygina, V.R, Vlaskina, A.V, Gabdulkhakov, A, Subach, O.M, Subach, F.V. | | Deposit date: | 2023-08-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 21, 2024
|
|
8QDD
 
 | | Structure of mBaoJin at pH 8.5 | | Descriptor: | CHLORIDE ION, SULFATE ION, mBaoJin | | Authors: | Samygina, V.R, Vlaskina, A.V, Gabdulkhakov, A, Subach, O.M, Subach, F.V. | | Deposit date: | 2023-08-28 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 21, 2024
|
|
8Q79
 
 | | Structure of mBaoJin at pH 6.5 | | Descriptor: | CHLORIDE ION, mBaoJin | | Authors: | Samygina, V.R, Subach, O.M, Vlaskina, A.V, Nikolaeva, A.Y, Borshchevsky, V, Qin, W, Subach, F.V. | | Deposit date: | 2023-08-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 21, 2024
|
|
3BC5
 
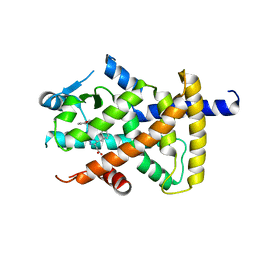 | | X-ray crystal structure of human ppar gamma with 2-(5-(3-(2-(5-methyl-2-phenyloxazol-4-yl)ethoxy)benzyl)-2-phenyl-2h-1,2,3-triazol-4-yl)acetic acid | | Descriptor: | (5-{3-[2-(5-methyl-2-phenyl-1,3-oxazol-4-yl)ethoxy]benzyl}-2-phenyl-2H-1,2,3-triazol-4-yl)acetic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2007-11-12 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Design, synthesis and structure-activity relationships of azole acids as novel, potent dual PPAR alpha/gamma agonists.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6XP5
 
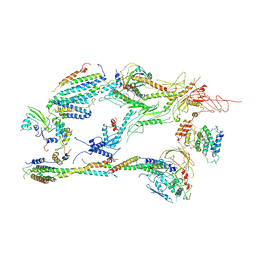 | | Head-Middle module of Mediator | | Descriptor: | HEAT, Med22, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Zhang, H.Q, Chen, D.C, Kornberg, R.D. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mediator structure and conformation change.
Mol.Cell, 81, 2021
|
|
6WNK
 
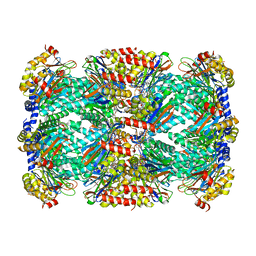 | | Macrocyclic peptides TDI5575 that selectively inhibit the Mycobacterium tuberculosis proteasome | | Descriptor: | (12S,15S)-N-[(2-fluorophenyl)methyl]-10,13-dioxo-12-{2-oxo-2-[(2R)-2-phenylpyrrolidin-1-yl]ethyl}-2-oxa-11,14-diazatricyclo[15.2.2.1~3,7~]docosa-1(19),3(22),4,6,17,20-hexaene-15-carboxamide, CITRIC ACID, DIMETHYLFORMAMIDE, ... | | Authors: | Hsu, H.C, Li, H. | | Deposit date: | 2020-04-22 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Macrocyclic Peptides that Selectively Inhibit the Mycobacterium tuberculosis Proteasome.
J.Med.Chem., 64, 2021
|
|
4KBQ
 
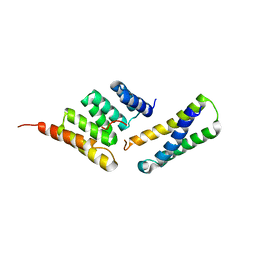 | | Structure of the CHIP-TPR domain in complex with the Hsc70 Lid-Tail domains | | Descriptor: | E3 ubiquitin-protein ligase CHIP, Heat shock cognate 71 kDa protein | | Authors: | Page, R.C, Amick, J, Nix, J.C, Misra, S. | | Deposit date: | 2013-04-23 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A Bipartite Interaction between Hsp70 and CHIP Regulates Ubiquitination of Chaperoned Client Proteins.
Structure, 23, 2015
|
|
6UCG
 
 | |
5YH3
 
 | | The structure of hFam20C and hFam20A complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Extracellular serine/threonine protein kinase FAM20C, Pseudokinase FAM20A | | Authors: | Zhu, Q, Xiao, J. | | Deposit date: | 2017-09-27 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and evolution of the Fam20 kinases
Nat Commun, 9, 2018
|
|
6JYV
 
 | | Structure of an isopenicillin N synthase from Pseudomonas aeruginosa PAO1 | | Descriptor: | Probable iron/ascorbate oxidoreductase, SODIUM ION | | Authors: | Hao, Z, Che, S, Wang, R, Liu, R, Zhang, Q, Bartlam, M. | | Deposit date: | 2019-04-28 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural characterization of an isopenicillin N synthase family oxygenase from Pseudomonas aeruginosa PAO1.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
8WFX
 
 | | Cryo-EM structure of CRISPR-Csm effector complex from Mycobacterium canettii | | Descriptor: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | Authors: | Huo, Y, Ma, X, Jiang, T. | | Deposit date: | 2023-09-20 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Type-III-A structure of mycobacteria CRISPR-Csm complexes involving atypical crRNAs.
Int.J.Biol.Macromol., 260, 2024
|
|
