8HIJ
 
 | | The 5-MTHF-bound BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | Anti-BRIL Fab heavy chain, Anti-BRIL Fab light chain, Anti-Fab nanobody, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
8HIK
 
 | | The TPP-bound BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | Anti-BRIL Fab heavy chain, Anti-BRIL Fab light chain, Anti-Fab nanobody, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
8HII
 
 | | The BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | BRIL-SLC19A1 chimera, anti-BRIL Fab heavy chain, anti-BRIL Fab light chain, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
8VHH
 
 | | Engineered holo tryptophan synthase (Tm9D8*) derived from T. maritima TrpB | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Porter, N.J, Johnston, K.E, Almhjell, P.J, Arnold, F.H. | | Deposit date: | 2024-01-02 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A combinatorially complete epistatic fitness landscape in an enzyme active site.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8HPB
 
 | |
8HQB
 
 | | NMR Structure of OsCIE1-Ubox | | Descriptor: | U-box domain-containing protein 12 | | Authors: | Zhang, Y, Yu, C.Z, Lan, W.X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Release of a ubiquitin brake activates OsCERK1-triggered immunity in rice.
Nature, 629, 2024
|
|
6JBI
 
 | | Structure of Tps1 apo structure | | Descriptor: | Trehalose-6-phosphate synthase | | Authors: | Wang, S, Zhao, Y, Yi, L, Wang, D, Liu, J. | | Deposit date: | 2019-01-25 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Magnaporthe oryzae trehalose-6-phosphate synthase (MoTps1) suggest a model for catalytic process of Tps1.
Biochem.J., 476, 2019
|
|
6JAK
 
 | | OtsA apo structure | | Descriptor: | Trehalose-6-phosphate synthase | | Authors: | Wang, S, Zhao, Y, Wang, D, Liu, J. | | Deposit date: | 2019-01-24 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structures of Magnaporthe oryzae trehalose-6-phosphate synthase (MoTps1) suggest a model for catalytic process of Tps1.
Biochem.J., 476, 2019
|
|
8HCI
 
 | |
8HI8
 
 | |
8HI7
 
 | |
6LDF
 
 | |
6LDE
 
 | |
6LDG
 
 | |
6JBR
 
 | | Tps1/UDP/T6P complex | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, Trehalose-6-phosphate synthase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, S, Zhao, Y, Wang, D, Liu, J. | | Deposit date: | 2019-01-26 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structures of Magnaporthe oryzae trehalose-6-phosphate synthase (MoTps1) suggest a model for catalytic process of Tps1.
Biochem.J., 476, 2019
|
|
6JGF
 
 | |
6JGV
 
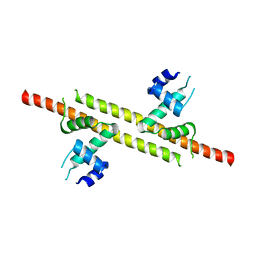 | |
6J1N
 
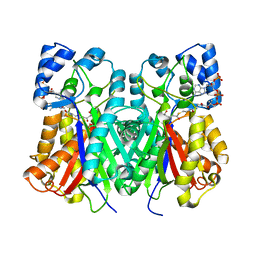 | | Anisodus acutangulus type III polyketide sythase AaPKS2 in complex with 4-carboxy-3-oxobutanoyl-CoA | | Descriptor: | (3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-10,14,19,21-tetraoxo-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphatricosan-23-oic acid 3,5-dioxide (non-preferred name), A. acutangulus PKS2 | | Authors: | Fang, C.L, Zhang, Y. | | Deposit date: | 2018-12-28 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Tropane alkaloids biosynthesis involves an unusual type III polyketide synthase and non-enzymatic condensation.
Nat Commun, 10, 2019
|
|
6JNI
 
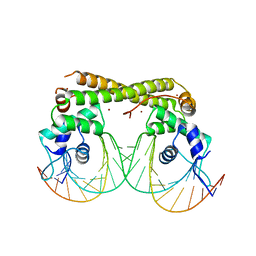 | |
6KC0
 
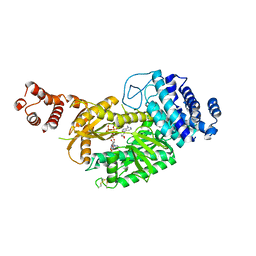 | | fused To-MtbCsm1 with 2ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A),CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), MAGNESIUM ION | | Authors: | Li, T, Huo, Y, Jiang, T. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Mycobacterium tuberculosis CRISPR/Cas system Csm1 holds clues to the evolutionary relationship between DNA polymerase and cyclase activity.
Int.J.Biol.Macromol., 170, 2020
|
|
8K14
 
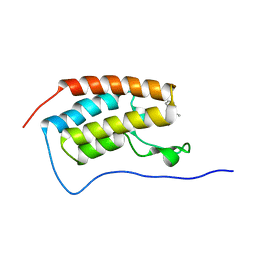 | | X-ray crystal structure of 18a in BRD4(1) | | Descriptor: | 4-[8-methoxy-2-methyl-1-(1-phenylethyl)imidazo[4,5-c]quinolin-7-yl]-3,5-dimethyl-1,2-oxazole, Bromodomain-containing protein 4 | | Authors: | Xu, H, Shen, H, Zhang, Y, Xu, Y, Li, R. | | Deposit date: | 2023-07-10 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Discovery of (R)-4-(8-methoxy-2-methyl-1-(1-phenylethy)-1H-imidazo[4,5-c]quinnolin-7-yl)-3,5-dimethylisoxazole as a potent and selective BET inhibitor for treatment of acute myeloid leukemia (AML) guided by FEP calculation.
Eur.J.Med.Chem., 263, 2024
|
|
6KTO
 
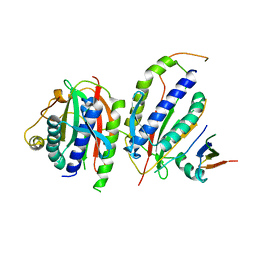 | | Crystal structure of human SHLD3-C-REV7-O-REV7-SHLD2 complex | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, Shieldin complex subunit 2, Shieldin complex subunit 3 | | Authors: | Liang, L, Yin, Y. | | Deposit date: | 2019-08-28 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4497633 Å) | | Cite: | Molecular basis for assembly of the shieldin complex and its implications for NHEJ.
Nat Commun, 11, 2020
|
|
8K31
 
 | | The complex of WRKY33 C terminal DBD and SIB1 | | Descriptor: | Probable WRKY transcription factor 33, Sigma factor binding protein 1, chloroplastic, ... | | Authors: | Dong, X, Gong, Z, Hu, Y.F. | | Deposit date: | 2023-07-14 | | Release date: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the regulation of plant transcription factor WRKY33 by the VQ protein SIB1.
Commun Biol, 7, 2024
|
|
8JAT
 
 | |
6JGX
 
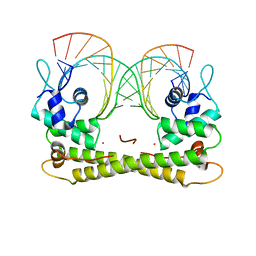 | | Crystal structure of the transcriptional regulator CadR from P. putida in complex with Cadmium(II) and DNA | | Descriptor: | CADMIUM ION, CadR, DNA (5'-D(*CP*AP*CP*CP*CP*TP*AP*TP*AP*GP*TP*GP*GP*CP*TP*AP*CP*AP*GP*GP*GP*T)-3'), ... | | Authors: | Liu, X.C, Gan, J.H, Chen, H. | | Deposit date: | 2019-02-15 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Selective cadmium regulation mediated by a cooperative binding mechanism in CadR.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
