5L6O
 
 | | EphB3 kinase domain covalently bound to an irreversible inhibitor (compound 3) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 1-(4-phenylazanylquinazolin-7-yl)ethanone, Ephrin type-B receptor 3 | | Authors: | Schimpl, M, Overman, R, Kung, A, Chen, Y.-C, Ni, F, Zhu, J, Turner, M, Molina, H, Zhang, C. | | Deposit date: | 2016-05-30 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Development of Specific, Irreversible Inhibitors for a Receptor Tyrosine Kinase EphB3.
J.Am.Chem.Soc., 138, 2016
|
|
1M07
 
 | | RESIDUES INVOLVED IN THE CATALYSIS AND BASE SPECIFICITY OF CYTOTOXIC RIBONUCLEASE FROM BULLFROG (RANA CATESBEIANA) | | Descriptor: | 5'-D(*AP*CP*GP*A)-3', Ribonuclease | | Authors: | Leu, Y.-J, Chern, S.-S, Wang, S.-C, Hsiao, Y.-Y, Amiraslanov, I, Liaw, Y.-C, Liao, Y.-D. | | Deposit date: | 2002-06-12 | | Release date: | 2003-01-21 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Residues involved in the catalysis, base specificity, and cytotoxicity of ribonuclease from Rana catesbeiana based upon mutagenesis and X-ray crystallography
J.Biol.Chem., 278, 2003
|
|
1BC4
 
 | | THE SOLUTION STRUCTURE OF A CYTOTOXIC RIBONUCLEASE FROM THE OOCYTES OF RANA CATESBEIANA (BULLFROG), NMR, 15 STRUCTURES | | Descriptor: | RIBONUCLEASE | | Authors: | Chang, C.-F, Chen, C, Chen, Y.-C, Hom, K, Huang, R.-F, Huang, T. | | Deposit date: | 1998-05-05 | | Release date: | 1998-10-14 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a cytotoxic ribonuclease from the oocytes of Rana catesbeiana (bullfrog).
J.Mol.Biol., 283, 1998
|
|
5JX5
 
 | | GH6 Orpinomyces sp. Y102 enzyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Tsai, L.C, Huang, H.C. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-17 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of GH6 Orpinomyces sp. Y102 CelC7 enzyme with exo- and endo- activity in a complex with cellobiose
Acta Crystallogr.,Sect.D, 2019
|
|
4QUC
 
 | | Crystal structure of chromodomain of Rhino | | Descriptor: | RE36324p | | Authors: | Li, S, Patel, D.J. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing.
Genes Dev., 28, 2014
|
|
4QUF
 
 | | crystal structure of chromodomain of Rhino with H3K9me3 | | Descriptor: | H3(1-15)K9me3 peptide, RE36324p | | Authors: | Li, S, Patel, D.J. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing.
Genes Dev., 28, 2014
|
|
7TYQ
 
 | | TEAD2 bound to Compound 1 | | Descriptor: | Transcriptional enhancer factor TEF-4, ethyl (8S)-7-oxo-5-[4-(trifluoromethyl)phenyl]-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carboxylate | | Authors: | Noland, C.L, Fong, R. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Novel mechanism of YAP-TEAD inhibition results in targeted chromatin remodeling and reveals an
expanded Hippo dependent landscape in cancers
To Be Published
|
|
7TYU
 
 | | TEAD2 bound to Compound 2 | | Descriptor: | (3R)-1-[(8S)-5-(4-cyclohexylphenyl)-7-oxo-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonyl]pyrrolidine-3-carbonitrile, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcriptional enhancer factor TEF-4 | | Authors: | Noland, C.L, Fong, R. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Novel mechanism of YAP-TEAD inhibition results in targeted chromatin remodeling and reveals an
expanded Hippo dependent landscape in cancers
To Be Published
|
|
7TYP
 
 | | TEAD2 bound to GNE-7883 | | Descriptor: | (8S)-5-(4-cyclohexylphenyl)-3-[3-(fluoromethyl)azetidine-1-carbonyl]-2-(3-methylpyrazin-2-yl)pyrazolo[1,5-a]pyrimidin-7(4H)-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcriptional enhancer factor TEF-4 | | Authors: | Noland, C.L, Fong, R. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel mechanism of YAP-TEAD inhibition results in targeted chromatin remodeling and reveals an
expanded Hippo dependent landscape in cancers
To Be Published
|
|
6OQO
 
 | | CDK6 in complex with Cpd24 N-(5-(6-ethyl-2,6-diazaspiro[3.3]heptan-2-yl)pyridin-2-yl)-5-fluoro-4-(4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl)pyrimidin-2-amine | | Descriptor: | Cyclin-dependent kinase 6, N-[5-(6-ethyl-2,6-diazaspiro[3.3]heptan-2-yl)pyridin-2-yl]-5-fluoro-4-[(4R)-4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl]pyrimidin-2-amine | | Authors: | Murray, J.M, Boenig, G.D.L. | | Deposit date: | 2019-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Design of a brain-penetrant CDK4/6 inhibitor for glioblastoma.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6OQL
 
 | | CDK6 in complex with Cpd13 (R)-5-fluoro-4-(4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl)-N-(5-(4-methylpiperazin-1-yl)pyridin-2-yl)pyrimidin-2-amine | | Descriptor: | 5-fluoro-N-[5-(4-methylpiperazin-1-yl)pyridin-2-yl]-4-[(4R)-4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl]pyrimidin-2-amine, Cyclin-dependent kinase 6 | | Authors: | Murray, J.M. | | Deposit date: | 2019-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.707 Å) | | Cite: | Design of a brain-penetrant CDK4/6 inhibitor for glioblastoma.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
7LVN
 
 | |
6CAD
 
 | | Crystal structure of RAF kinase domain bound to the inhibitor 2a | | Descriptor: | 1-(propan-2-yl)-3-({3-[3-(trifluoromethyl)phenyl]isoquinolin-8-yl}ethynyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Serine/threonine-protein kinase B-raf | | Authors: | Maisonneuve, P, Kurinov, I, Assadieskandar, A, Yu, C, Liu, X, Chen, Y.-C, Prakash, G.K.S, Zhang, C, SIcheri, F. | | Deposit date: | 2018-01-30 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Effects of rigidity on the selectivity of protein kinase inhibitors.
Eur J Med Chem, 146, 2018
|
|
5W0T
 
 | |
2M0J
 
 | |
2M0K
 
 | |
7LTN
 
 | | Crystal structure of Mpro in complex with inhibitor CDD-1713 | | Descriptor: | 2-[4-(1~{H}-indazol-4-yl)-2-methanoyl-6-methoxy-phenoxy]-~{N},~{N}-dimethyl-ethanamide, 3C-like proteinase | | Authors: | Lu, S, Palzkill, T, Matzuk, M, Young, D, Melek, N, Chamakuri, S. | | Deposit date: | 2021-02-19 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | DNA-encoded chemistry technology yields expedient access to SARS-CoV-2 M pro inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EQV
 
 | | Crystal structure of JMJD2A complexed with 3,4-dihydroxybenzoic acid | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, CHLORIDE ION, Lysine-specific demethylase 4A, ... | | Authors: | Fang, W.-K, Yang, S.-M, Wang, W.-C. | | Deposit date: | 2021-05-04 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Natural product myricetin is a pan-KDM4 inhibitor which with poly lactic-co-glycolic acid formulation effectively targets castration-resistant prostate cancer.
J.Biomed.Sci., 29, 2022
|
|
6OQI
 
 | | CDK2 in complex with Cpd14 (5-fluoro-4-(4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl)-N-(5-(4-methylpiperazin-1-yl)pyridin-2-yl)pyrimidin-2-amine) | | Descriptor: | 5-fluoro-N-[5-(4-methylpiperazin-1-yl)pyridin-2-yl]-4-[(4S)-4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl]pyrimidin-2-amine, Cyclin-dependent kinase 2 | | Authors: | Murray, J.M. | | Deposit date: | 2019-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of a brain-penetrant CDK4/6 inhibitor for glioblastoma.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
8U89
 
 | | The structure of the PP2A-B56Delta holoenzyme mutant - E197K | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit delta isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Wu, C.G, Xing, Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | B56 delta long-disordered arms form a dynamic PP2A regulation interface coupled with global allostery and Jordan's syndrome mutations.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U1X
 
 | | The structure of the PP2A-B56Delta holoenzyme mutant - E197K | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit delta isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Wu, C.G, Xing, Y. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | B56 delta long-disordered arms form a dynamic PP2A regulation interface coupled with global allostery and Jordan's syndrome mutations.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6IDW
 
 | | GH6 Orpinomyces sp. Y102 enzyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Tsai, L.C, Huang, H.C. | | Deposit date: | 2018-09-11 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structures of the GH6 Orpinomyces sp. Y102 CelC7 enzyme with exo and endo activity and its complex with cellobiose.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6UCH
 
 | | SMARCB1 nucleosome-interacting C-terminal alpha helix | | Descriptor: | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Valencia, A.M, Sun, Z.Y.J, Seo, H.S, Vangos, H.S, Yeoh, Z.C, Mashtalir, N, Dhe-Paganon, S, Kadoch, C. | | Deposit date: | 2019-09-16 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recurrent SMARCB1 Mutations Reveal a Nucleosome Acidic Patch Interaction Site That Potentiates mSWI/SNF Complex Chromatin Remodeling.
Cell, 179, 2019
|
|
7VUE
 
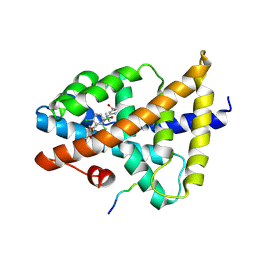 | | Structural insight of the molecular mechanism of cilofexor bound to FXR | | Descriptor: | 2-[3-[4-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-2-chloranyl-phenyl]-3-oxidanyl-azetidin-1-yl]pyridine-4-carboxylic acid, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | Authors: | Jiang, L, Chen, Y.C. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural insight into the molecular mechanism of cilofexor binding to the farnesoid X receptor.
Biochem.Biophys.Res.Commun., 595, 2022
|
|
7UR9
 
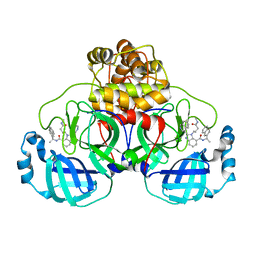 | | SARS-Cov2 Main protease in complex with inhibitor CDD-1845 | | Descriptor: | (2P)-2-(isoquinolin-4-yl)-1-[4-(methylamino)-4-oxobutyl]-N-[(1S)-1-(naphthalen-2-yl)ethyl]-1H-benzimidazole-7-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lu, S, Palzkill, T. | | Deposit date: | 2022-04-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | DNA-encoded chemical libraries yield non-covalent and non-peptidic SARS-CoV-2 main protease inhibitors.
Commun Chem, 6, 2023
|
|
