1DPR
 
 | | STRUCTURES OF THE APO-AND METAL ION ACTIVATED FORMS OF THE DIPHTHERIA TOX REPRESSOR FROM CORYNEBACTERIUM DIPHTHERIAE | | Descriptor: | DIPHTHERIA TOX REPRESSOR | | Authors: | Schiering, N, Tao, X, Murphy, J, Petsko, G.A, Ringe, D. | | Deposit date: | 1995-02-06 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the apo- and the metal ion-activated forms of the diphtheria tox repressor from Corynebacterium diphtheriae.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
6XIS
 
 | |
6XIT
 
 | | Cryo-EM structure of the G protein-gated inward rectifier K+ channel GIRK2 (Kir3.2) in complex with PIP2 | | Descriptor: | G protein-activated inward rectifier potassium channel 2, POTASSIUM ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Niu, Y, Tao, X, MacKinnon, R. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM analysis of PIP 2 regulation in mammalian GIRK channels.
Elife, 9, 2020
|
|
5BXJ
 
 | | Complex of the Fk1 domain mutant A19T of FKBP51 with 4-Nitrophenol | | Descriptor: | P-NITROPHENOL, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wu, D, Tao, X, Chen, Z, Han, J, Jia, W, Li, X, Wang, Z, He, Y.X. | | Deposit date: | 2015-06-09 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | The environmental endocrine disruptor p-nitrophenol interacts with FKBP51, a positive regulator of androgen receptor and inhibits androgen receptor signaling in human cells
J. Hazard. Mater., 307, 2016
|
|
1FYV
 
 | | CRYSTAL STRUCTURE OF THE TIR DOMAIN OF HUMAN TLR1 | | Descriptor: | TOLL-LIKE RECEPTOR 1 | | Authors: | Xu, Y, Tao, X, Shen, B, Horng, T, Medzhitov, R, Manley, J.L, Tong, L. | | Deposit date: | 2000-10-03 | | Release date: | 2000-11-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for signal transduction by the Toll/interleukin-1 receptor domains.
Nature, 408, 2000
|
|
1FYX
 
 | | CRYSTAL STRUCTURE OF P681H MUTANT OF TIR DOMAIN OF HUMAN TLR2 | | Descriptor: | TOLL-LIKE RECEPTOR 2 | | Authors: | Xu, Y, Tao, X, Shen, B, Horng, T, Medzhitov, R, Manley, J.L, Tong, L. | | Deposit date: | 2000-10-03 | | Release date: | 2000-11-22 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for signal transduction by the Toll/interleukin-1 receptor domains.
Nature, 408, 2000
|
|
1FYW
 
 | | CRYSTAL STRUCTURE OF THE TIR DOMAIN OF HUMAN TLR2 | | Descriptor: | TOLL-LIKE RECEPTOR 2 | | Authors: | Xu, Y, Tao, X, Shen, B, Horng, T, Medzhitov, R, Manley, J.L, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-10-03 | | Release date: | 2000-11-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for signal transduction by the Toll/interleukin-1 receptor domains.
Nature, 408, 2000
|
|
2GVL
 
 | | Crystal Structure of Murine NMPRTase | | Descriptor: | Nicotinamide phosphoribosyltransferase | | Authors: | Khan, J.A, Tao, X, Tong, L. | | Deposit date: | 2006-05-02 | | Release date: | 2006-07-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the inhibition of human NMPRTase, a novel target for anticancer agents.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2GVJ
 
 | | Crystal Structure of Human NMPRTase in complex with FK866 | | Descriptor: | (2E)-N-{4-[1-(benzenecarbonyl)piperidin-4-yl]butyl}-3-(pyridin-3-yl)prop-2-enamide, Nicotinamide phosphoribosyltransferase | | Authors: | Khan, J.A, Tao, X, Tong, L. | | Deposit date: | 2006-05-02 | | Release date: | 2006-06-20 | | Last modified: | 2016-11-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the inhibition of human NMPRTase, a novel target for anticancer agents.
Nat.Struct.Mol.Biol., 13, 2006
|
|
5TJI
 
 | | Ca2+ bound aplysia Slo1 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, High conductance calcium-activated potassium channel | | Authors: | MacKinnon, R, Tao, X, Hite, R.K. | | Deposit date: | 2016-10-04 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for gating the high-conductance Ca(2+)-activated K(+) channel.
Nature, 541, 2017
|
|
2GVG
 
 | | Crystal Structure of human NMPRTase and its complex with NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Khan, J.A, Tao, X, Tong, L. | | Deposit date: | 2006-05-02 | | Release date: | 2006-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for the inhibition of human NMPRTase, a novel target for anticancer agents.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2JS7
 
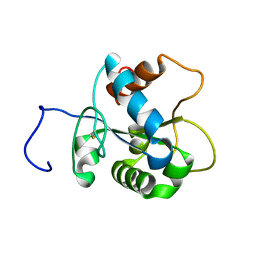 | | Solution NMR structure of human myeloid differentiation primary response (MyD88). Northeast Structural Genomics target HR2869A | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Rossi, P, Ramelot, T.A, Tao, X, Ciano, M, Ho, C, Ma, L.-C, Xiao, R, Acton, T.B, Kennedy, M.A, Tong, L, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-10-23 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Human Myeloid Differentiation Primary Response (MyD88).
To be Published
|
|
3SPH
 
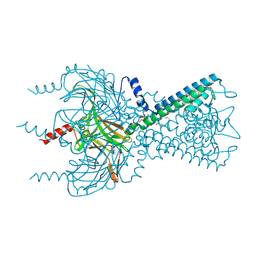 | | Inward rectifier potassium channel Kir2.2 I223L mutant in complex with PIP2 | | Descriptor: | Inward-rectifier K+ channel Kir2.2, POTASSIUM ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Hansen, S.B, Tao, X, MacKinnon, R. | | Deposit date: | 2011-07-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural basis of PIP(2) activation of the classical inward rectifier K(+) channel Kir2.2.
Nature, 477, 2011
|
|
3SPJ
 
 | |
3SPC
 
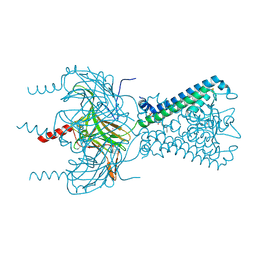 | | Inward rectifier potassium channel Kir2.2 in complex with dioctanoylglycerol pyrophosphate (DGPP) | | Descriptor: | (2R)-3-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}propane-1,2-diyl dioctanoate, Inward-rectifier K+ channel Kir2.2, POTASSIUM ION | | Authors: | Hansen, S.B, Tao, X, MacKinnon, R. | | Deposit date: | 2011-07-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.454 Å) | | Cite: | Structural basis of PIP(2) activation of the classical inward rectifier K(+) channel Kir2.2.
Nature, 477, 2011
|
|
3SPG
 
 | | Inward rectifier potassium channel Kir2.2 R186A mutant in complex with PIP2 | | Descriptor: | Inward-rectifier K+ channel Kir2.2, POTASSIUM ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Hansen, S.B, Tao, X, MacKinnon, R. | | Deposit date: | 2011-07-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | Structural basis of PIP(2) activation of the classical inward rectifier K(+) channel Kir2.2.
Nature, 477, 2011
|
|
3SPI
 
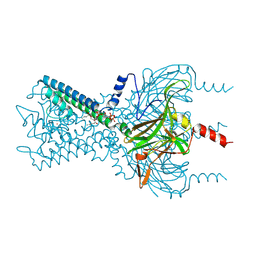 | | Inward rectifier potassium channel Kir2.2 in complex with PIP2 | | Descriptor: | Inward-rectifier K+ channel Kir2.2, POTASSIUM ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Hansen, S.B, Tao, X, MacKinnon, R. | | Deposit date: | 2011-07-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.307 Å) | | Cite: | Structural basis of PIP(2) activation of the classical inward rectifier K(+) channel Kir2.2.
Nature, 477, 2011
|
|
7N0L
 
 | |
7N0K
 
 | |
8X8M
 
 | | Cryo-EM structure of a bacteriophage tail- spike protein against Klebsiella pneumoniae K64,ORF41(K64-ORF41) | | Descriptor: | Probable tail spike protein | | Authors: | Xie, Y, Huang, T, Zhang, Z, Tao, X. | | Deposit date: | 2023-11-27 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Structural and functional basis of bacteriophage K64-ORF41 depolymerase for capsular polysaccharide degradation of Klebsiella pneumoniae K64.
Int.J.Biol.Macromol., 265, 2024
|
|
8X8O
 
 | | Cryo-EM structure of a bacteriophage tail- spike protein against Klebsiella pneumoniae K64,ORF41(K64-ORF41) in 5 mM EDTA | | Descriptor: | Probable tail spike protein | | Authors: | Xie, Y, Huang, T, Zhang, Z, Tao, X. | | Deposit date: | 2023-11-27 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural and functional basis of bacteriophage K64-ORF41 depolymerase for capsular polysaccharide degradation of Klebsiella pneumoniae K64.
Int.J.Biol.Macromol., 265, 2024
|
|
5Y28
 
 | | Crystal structure of H. pylori HtrA with PDZ2 deletion | | Descriptor: | 1,2-ETHANEDIOL, Periplasmic serine endoprotease DegP-like, UNK-UNK-UNK-UNK | | Authors: | Zhang, Z, Huang, Q, Tao, X. | | Deposit date: | 2017-07-24 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.08606815 Å) | | Cite: | The unique trimeric assembly of the virulence factor HtrA fromHelicobacter pylorioccurs via N-terminal domain swapping.
J.Biol.Chem., 294, 2019
|
|
5Y2D
 
 | | Crystal structure of H. pylori HtrA | | Descriptor: | Periplasmic serine endoprotease DegP-like, UNK-UNK-K-UNK-UNK-UNK-UNK-UNK-UNK-UNK, UNK-UNK-UNK, ... | | Authors: | Zhang, Z, Huang, Q, Tao, X. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.70009851 Å) | | Cite: | The unique trimeric assembly of the virulence factor HtrA fromHelicobacter pylorioccurs via N-terminal domain swapping.
J.Biol.Chem., 294, 2019
|
|
8GQD
 
 | | Complex Structure of Arginine Kinase McsB and McsA from Staphylococcus aureus | | Descriptor: | Protein-arginine kinase, Protein-arginine kinase activator protein, ZINC ION | | Authors: | Lu, K, Luo, B, Tao, X, Li, H, Xie, Y, Zhao, Z, Xia, W, Su, Z, Mao, Z. | | Deposit date: | 2022-08-30 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Complex structure and activation mechanism of arginine kinase McsB by McsA.
Nat.Chem.Biol., 2024
|
|
7CX4
 
 | | Cryo-EM structure of the Evatanepag-bound EP2-Gs complex | | Descriptor: | 2-[3-[[(4-~{tert}-butylphenyl)methyl-pyridin-3-ylsulfonyl-amino]methyl]phenoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
