3RGX
 
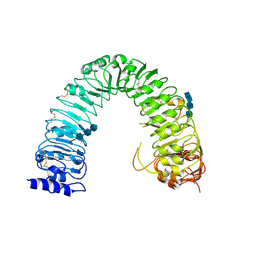 | | Structural insight into brassinosteroid perception by BRI1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chai, J, Han, Z, She, J, Wang, J, Cheng, W, Wang, J. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural insight into brassinosteroid perception by BRI1.
Nature, 474, 2011
|
|
3RGZ
 
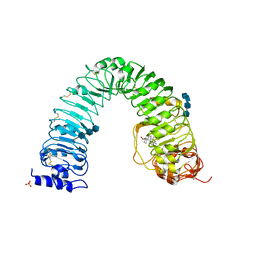 | | Structural insight into brassinosteroid perception by BRI1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chai, J, Han, Z, She, J, Wang, J, Cheng, W. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Structural insight into brassinosteroid perception by BRI1.
Nature, 474, 2011
|
|
1L5B
 
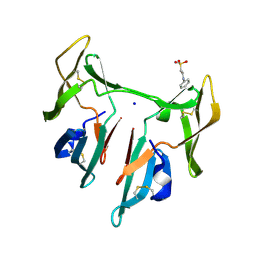 | | DOMAIN-SWAPPED CYANOVIRIN-N DIMER | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, SODIUM ION, cyanovirin-N | | Authors: | Barrientos, L.G, Louis, J.M, Botos, I, Mori, T, Han, Z, O'Keefe, B.R, Boyd, M.R, Wlodawer, A, Gronenborn, A.M. | | Deposit date: | 2002-03-06 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The domain-swapped dimer of cyanovirin-N is in a metastable folded state: reconciliation of X-ray and NMR structures.
Structure, 10, 2002
|
|
1L5E
 
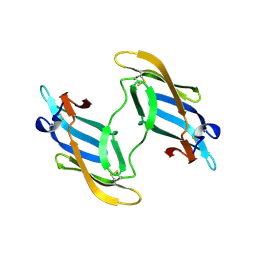 | | The domain-swapped dimer of CV-N in solution | | Descriptor: | Cyanovirin-N | | Authors: | Barrientos, L.G, Louis, J.M, Botos, I, Mori, T, Han, Z, O'Keefe, B.R, Boyd, M.R, Wlodawer, A, Gronenborn, A.M. | | Deposit date: | 2002-03-06 | | Release date: | 2002-06-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The domain-swapped dimer of cyanovirin-N is in a metastable folded state: reconciliation of X-ray and NMR structures.
Structure, 10, 2002
|
|
4L6R
 
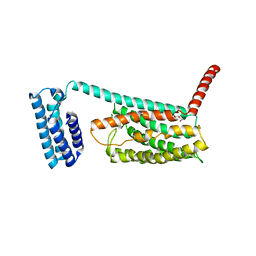 | | Structure of the class B human glucagon G protein coupled receptor | | Descriptor: | DI(HYDROXYETHYL)ETHER, Soluble cytochrome b562 and Glucagon receptor chimera | | Authors: | Siu, F.Y, He, M, de Graaf, C, Han, G.W, Yang, D, Zhang, Z, Zhou, C, Xu, Q, Wacker, D, Joseph, J.S, Liu, W, Lau, J, Cherezov, V, Katritch, V, Wang, M.W, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2013-06-12 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the human glucagon class B G-protein-coupled receptor.
Nature, 499, 2013
|
|
4HPZ
 
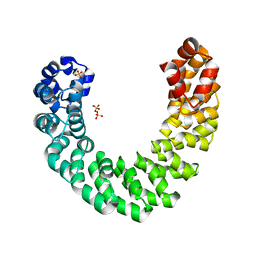 | |
7O5N
 
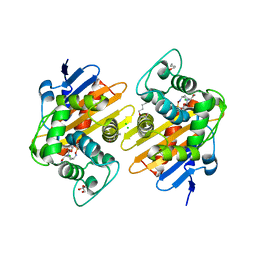 | | Crystal Structure of a Class D carbapenemase complexed with Avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1-BUTANOL, Beta-lactamase, ... | | Authors: | Zhou, Q, Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-04-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Ion-Pair Induced Intermediate Complex Captured in Class D Carbapenemase Reveals Chloride Ion as a Janus Effector Modulating Activity
Acs Cent.Sci., 2023
|
|
6QLE
 
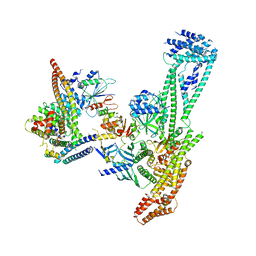 | | Structure of inner kinetochore CCAN complex | | Descriptor: | Central kinetochore subunit CTF3,Inner kinetochore subunit CTF3,Central kinetochore subunit CTF3,Inner kinetochore subunit CTF3, Central kinetochore subunit MCM16,Central kinetochore subunit MCM16,Inner kinetochore subunit MCM16,Mcm16p, Inner kinetochore subunit AME1,Inner kinetochore subunit AME1,Inner kinetochore subunit AME1,Inner kinetochore subunit AME1, ... | | Authors: | Yan, K, Yang, J, Zhang, Z, McLaughlin, S.H, Chang, L, Fasci, D, Heck, A.J.R, Barford, D. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Nature, 574, 2019
|
|
6QLD
 
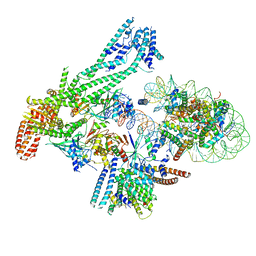 | | Structure of inner kinetochore CCAN-Cenp-A complex | | Descriptor: | DNA (125-MER), Histone H2A.1, Histone H2B.1, ... | | Authors: | Yan, K, Yang, J, Zhang, Z, McLaughlin, S.H, Chang, L, Fasci, D, Heck, A.J.R, Barford, D. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Nature, 574, 2019
|
|
6QLF
 
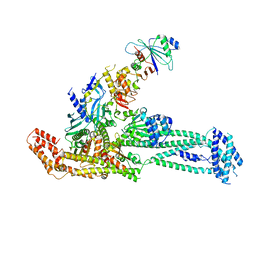 | | Structure of inner kinetochore CCAN complex with mask1 | | Descriptor: | Inner kinetochore subunit AME1, Inner kinetochore subunit CHL4, Inner kinetochore subunit CTF19, ... | | Authors: | Yan, K, Yang, J, Zhang, Z, McLaughlin, S.H, Chang, L, Fasci, D, Heck, A.J.R, Barford, D. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Nature, 574, 2019
|
|
4J0M
 
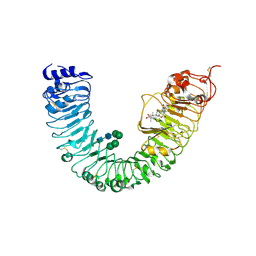 | | Crystal structure of BRL1 (LRR) in complex with brassinolide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Brassinolide, ... | | Authors: | Chai, J, She, J, Han, Z, Zhou, B. | | Deposit date: | 2013-01-31 | | Release date: | 2013-07-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for differential recognition of brassinolide by its receptors
Protein Cell, 4, 2013
|
|
7ON1
 
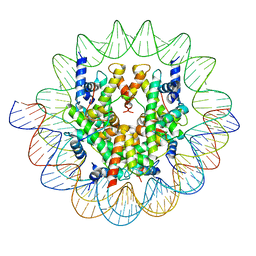 | | Cenp-A nucleosome in complex with Cenp-C | | Descriptor: | BJ4_G0006610.mRNA.1.CDS.1, BJ4_G0007000.mRNA.1.CDS.1, DNA (123-MER), ... | | Authors: | Yan, K, Yang, J, Zhang, Z, Barford, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cenp-A nucleosome in complex with Cenp-C
To Be Published
|
|
5MZ6
 
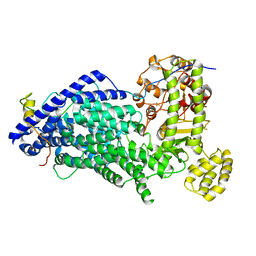 | | Cryo-EM structure of a Separase-Securin complex from Caenorhabditis elegans at 3.8 A resolution | | Descriptor: | Interactor of FizzY protein, SEParase | | Authors: | Boland, A, Martin, T.G, Zhang, Z, Yang, J, Bai, X.C, Chang, L, Scheres, S.H.W, Barford, D. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-08 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of a metazoan separase-securin complex at near-atomic resolution.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5LCW
 
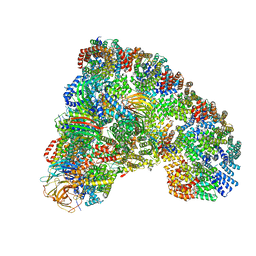 | | Cryo-EM structure of the Anaphase-promoting complex/Cyclosome, in complex with the Mitotic checkpoint complex (APC/C-MCC) at 4.2 angstrom resolution | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Alfieri, C, Chang, L, Zhang, Z, Yang, J, Maslen, S, Skehel, M, Barford, D. | | Deposit date: | 2016-06-22 | | Release date: | 2016-08-10 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular basis of APC/C regulation by the spindle assembly checkpoint.
Nature, 536, 2016
|
|
4MNA
 
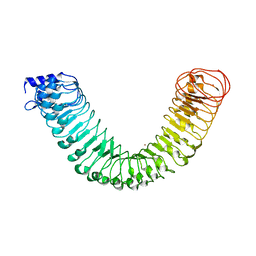 | | Crystal structure of the free FLS2 ectodomains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LRR receptor-like serine/threonine-protein kinase FLS2, ZINC ION | | Authors: | Chai, J, Han, Z, Sun, Y. | | Deposit date: | 2013-09-10 | | Release date: | 2013-12-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.998 Å) | | Cite: | Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex.
Science, 342, 2013
|
|
4MN8
 
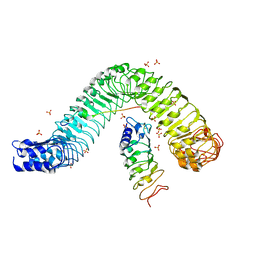 | | Crystal structure of flg22 in complex with the FLS2 and BAK1 ectodomains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1, ... | | Authors: | Chai, J, Sun, Y, Han, Z. | | Deposit date: | 2013-09-10 | | Release date: | 2013-12-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.062 Å) | | Cite: | Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex.
Science, 342, 2013
|
|
8HAY
 
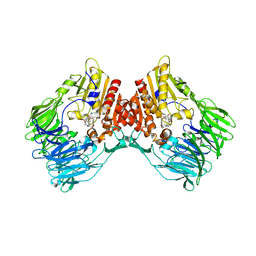 | | d4-bound btDPP4 | | Descriptor: | (1~{R})-1-[[4-[5-[[(1~{R})-6,7-dimethoxy-2-methyl-3,4-dihydro-1~{H}-isoquinolin-1-yl]methyl]-2-methoxy-phenoxy]phenyl]methyl]-6,7-dimethoxy-2-methyl-3,4-dihydro-1~{H}-isoquinoline, btDPP4 | | Authors: | Hang, J, Jiang, C, Wang, K, Zhang, Z, Guo, F, Liu, J, Wang, G, Lei, X, Gonzalez, F, Qiao, J. | | Deposit date: | 2022-10-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target.
Science, 381, 2023
|
|
4M7E
 
 | | Structural insight into BL-induced activation of the BRI1-BAK1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1, ... | | Authors: | Chai, J, Han, Z, Sun, Y. | | Deposit date: | 2013-08-12 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.602 Å) | | Cite: | Structural insight into BL-induced activation of the BRI1-BAK1 complex
To be Published
|
|
5VF3
 
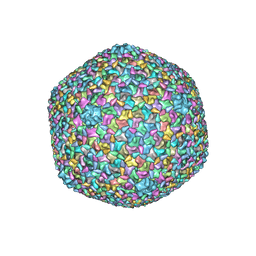 | | Bacteriophage T4 isometric capsid | | Descriptor: | Capsid vertex protein gp24, Highly immunogenic outer capsid protein, Major capsid protein, ... | | Authors: | Chen, Z, Sun, L, Zhang, Z, Fokine, A, Padilla-Sanchez, V, Hanein, D, Jiang, W, Rossmann, M.G, Rao, V.B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the bacteriophage T4 isometric head at 3.3- angstrom resolution and its relevance to the assembly of icosahedral viruses.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6TGC
 
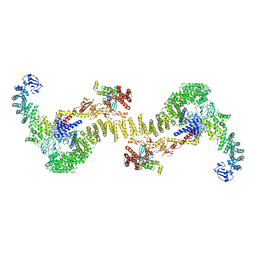 | | CryoEM structure of the ternary DOCK2-ELMO1-RAC1 complex. | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Chang, L, Yang, J, Chang, J.H, Zhang, Z, Boland, A, McLaughlin, S.H, Abu-Thuraia, A, Killoran, R.C, Smith, M.J, Cote, J.F, Barford, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
7SIM
 
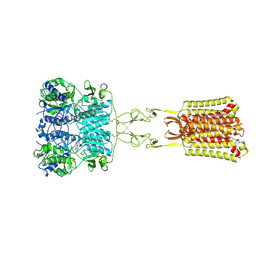 | | Structure of positive allosteric modulator-free active human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SIN
 
 | | Structure of negative allosteric modulator-bound inactive human calcium-sensing receptor | | Descriptor: | 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile, Isoform 1 of Extracellular calcium-sensing receptor | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SIL
 
 | | Structure of positive allosteric modulator-bound active human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-chlorophenyl)-N-[(1R)-1-(3-methoxyphenyl)ethyl]propan-1-amine, ... | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6TGB
 
 | | CryoEM structure of the binary DOCK2-ELMO1 complex | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1 | | Authors: | Chang, L, Yang, J, Chang, J.H, Zhang, Z, Boland, A, McLaughlin, S.H, Abu-Thuraia, A, Killoran, R.C, Smith, M.J, Cote, J.F, Barford, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
5BPT
 
 | |
