6PWS
 
 | |
6PWT
 
 | |
7QB1
 
 | | PPARg in complex with inhibitor | | Descriptor: | 3-[(3,4-dichlorophenyl)methyl]-4-oxidanylidene-6-phenoxy-phthalazine-1-carboxylic acid, Peroxisome proliferator-activated receptor gamma, SULFATE ION | | Authors: | Petersen, J. | | Deposit date: | 2021-11-17 | | Release date: | 2022-05-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery by Virtual Screening of an Inhibitor of CDK5-Mediated PPAR gamma Phosphorylation.
Acs Med.Chem.Lett., 13, 2022
|
|
6PWR
 
 | |
5KNZ
 
 | | Human Islet Amyloid Polypeptide Segment 19-SGNNFGAILSS-29 with Early Onset S20G Mutation Determined by MicroED | | Descriptor: | hIAPP(residues 19-29)S20G | | Authors: | Krotee, P.A.L, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Shi, D, Nannenga, B.L, Hattne, J, Reyes, F.E, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2016-06-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.9 Å) | | Cite: | Atomic structures of fibrillar segments of hIAPP suggest tightly mated beta-sheets are important for cytotoxicity.
Elife, 6, 2017
|
|
2RKX
 
 | | The 3D structure of chain D, cyclase subunit of imidazoleglycerol_evolvedcerolphosphate synthase | | Descriptor: | Cyclase subunit of imidazoleglycerol_evolvedcerolphosphate synthase | | Authors: | Tawfik, D, Khersonsky, O, Albeck, S, Dym, O, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2007-10-18 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Kemp elimination catalysts by computational enzyme design.
Nature, 453, 2008
|
|
5Z65
 
 | | Crystal structure of porcine aminopeptidase N ectodomain in functional form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, Y.G, Li, R, Jiang, L.G, Qiao, S.L, Zhang, G.P. | | Deposit date: | 2018-01-22 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Characterization of the interaction between recombinant porcine aminopeptidase N and spike glycoprotein of porcine epidemic diarrhea virus.
Int. J. Biol. Macromol., 117, 2018
|
|
5E4K
 
 | | Structure of ligand binding region of uPARAP at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, C-type mannose receptor 2, ... | | Authors: | Yuan, C, Huang, M. | | Deposit date: | 2015-10-06 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structures of the ligand-binding region of uPARAP: effect of calcium ion binding
Biochem.J., 473, 2016
|
|
6A8G
 
 | | The crystal structure of muPAin-1-IG in complex with muPA-SPD at pH8.5 | | Descriptor: | PHOSPHATE ION, Urokinase-type plasminogen activator chain B, muPAin-1-IG | | Authors: | Wang, D, Yang, Y.S, Jiang, L.G, Huang, M.D, Li, J.Y, Andreasen, P.A, Xu, P, Chen, Z. | | Deposit date: | 2018-07-08 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Suppression of Tumor Growth and Metastases by Targeted Intervention in Urokinase Activity with Cyclic Peptides.
J.Med.Chem., 62, 2019
|
|
6A8N
 
 | | The crystal structure of muPAin-1-IG-2 in complex with muPA-SPD at pH8.5 | | Descriptor: | CYS-PRO-ALA-TYR-SER-ARG-TYR-ILE-GLY-CYS, Urokinase-type plasminogen activator B | | Authors: | Wang, D, Yang, Y.S, Jiang, L.G, Huang, M.D, Li, J.Y, Andreasen, P.A, Xu, P, Chen, Z. | | Deposit date: | 2018-07-09 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | Suppression of Tumor Growth and Metastases by Targeted Intervention in Urokinase Activity with Cyclic Peptides.
J.Med.Chem., 62, 2019
|
|
4E0L
 
 | | FYLLYYT segment from human Beta 2 Microglobulin (62-68) displayed on 54-membered macrocycle scaffold | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclic pseudo-peptide FYLLYYT(ORN)KN(HAO)SA(ORN) | | Authors: | Zhao, M, Liu, C, Michael, S.R, Eisenberg, D. | | Deposit date: | 2012-03-04 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Out-of-register beta-sheets suggest a pathway to toxic amyloid aggregates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8ITR
 
 | |
8ITT
 
 | |
4XXB
 
 | | Crystal structure of human MDM2-RPL11 | | Descriptor: | 60S ribosomal protein L11, BETA-MERCAPTOETHANOL, E3 ubiquitin-protein ligase Mdm2, ... | | Authors: | Zheng, J, Chen, Z. | | Deposit date: | 2015-01-30 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human MDM2 complexed with RPL11 reveals the molecular basis of p53 activation
Genes Dev., 29, 2015
|
|
4ZSZ
 
 | |
4ZSV
 
 | |
4ZSX
 
 | |
5EW6
 
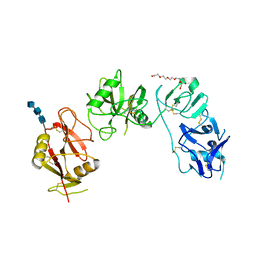 | | Structure of ligand binding region of uPARAP at pH 7.4 without calcium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type mannose receptor 2, ... | | Authors: | Yuan, C, Huang, M. | | Deposit date: | 2015-11-20 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structures of the ligand-binding region of uPARAP: effect of calcium ion binding
Biochem.J., 473, 2016
|
|
6AZR
 
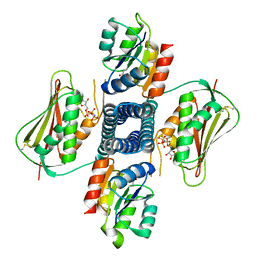 | | Crystal structure of the T264A HK853cp-BeF3-RR468 complex | | Descriptor: | Chemotaxis regulator-transmits chemoreceptor signals to flagelllar motor components CheY, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Rose, J, Zhou, P. | | Deposit date: | 2017-09-11 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.628 Å) | | Cite: | A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases.
Nat Commun, 8, 2017
|
|
6BCS
 
 | | LilrB2 D1D2 domains complexed with benzamidine | | Descriptor: | BENZAMIDINE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Cao, Q, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2017-10-20 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibiting amyloid-beta cytotoxicity through its interaction with the cell surface receptor LilrB2 by structure-based design.
Nat Chem, 10, 2018
|
|
8VDJ
 
 | | Crystal structure of SARS-CoV-2 3CL protease (3CLpro) as a covalent complex with EDP-235 | | Descriptor: | 3C-like proteinase nsp5, 4,6,7-trifluoro-N-{(2S)-1-[(3R,5'R)-5'-(iminomethyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidin]-1'-yl]-4-methyl-1-oxopentan-2-yl}-N-methyl-1H-indole-2-carboxamide, THIOCYANATE ION | | Authors: | Cade, I.A, Rhodin, M.H.J. | | Deposit date: | 2023-12-15 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | The small molecule inhibitor of SARS-CoV-2 3CLpro EDP-235 prevents viral replication and transmission in vivo.
Nat Commun, 15, 2024
|
|
8Y6F
 
 | |
7D5O
 
 | | C-Src in complex with TAS-120 | | Descriptor: | 1-[(3S)-3-{4-amino-3-[(3,5-dimethoxyphenyl)ethynyl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}pyrrolidin-1-yl]prop-2-en-1-one, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Qu, L.Z, Chen, Y.H. | | Deposit date: | 2020-09-27 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural insights into the potency and selectivity of covalent pan-FGFR inhibitors
Commun Chem, 5, 2022
|
|
7DHT
 
 | |
3OVJ
 
 | |
