7YHI
 
 | |
8H66
 
 | |
8H6D
 
 | |
8H6C
 
 | |
8H65
 
 | |
8HF5
 
 | |
7DPX
 
 | |
8HF6
 
 | | Cryo-EM structure of nucleotide-bound ComA E647Q mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Competence factor transporting ATP-binding protein/permease ComA | | Authors: | Yu, L, Xin, X, Min, L. | | Deposit date: | 2022-11-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of peptide secretion for Quorum sensing by ComA.
Nat Commun, 14, 2023
|
|
8HF4
 
 | |
7D7F
 
 | | Structure of PKD1L3-CTD/PKD2L1 in calcium-bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Polycystic kidney disease 2-like 1 protein, ... | | Authors: | Su, Q, Shi, Y.G. | | Deposit date: | 2020-10-03 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for Ca 2+ activation of the heteromeric PKD1L3/PKD2L1 channel.
Nat Commun, 12, 2021
|
|
8HF7
 
 | |
7YMS
 
 | |
1CF5
 
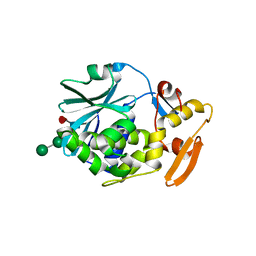 | | BETA-MOMORCHARIN STRUCTURE AT 2.55 A | | Descriptor: | PROTEIN (BETA-MOMORCHARIN), beta-D-xylopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yuan, Y.-R, He, Y.-N, Xiong, J.-P, Xia, Z.-X. | | Deposit date: | 1999-03-24 | | Release date: | 1999-06-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Three-dimensional structure of beta-momorcharin at 2.55 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
7YRH
 
 | |
7YRF
 
 | |
7YV7
 
 | |
7YV2
 
 | |
7YT9
 
 | | crystal structure of AGD1-4 of Arabidopsis AGDP3 | | Descriptor: | AGD1-4 of Arabidopsis AGDP3 | | Authors: | Zhou, X, Du, J. | | Deposit date: | 2022-08-13 | | Release date: | 2022-10-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The H3K9me2-binding protein AGDP3 limits DNA methylation and transcriptional gene silencing in Arabidopsis.
J Integr Plant Biol, 64, 2022
|
|
7YTA
 
 | |
6M2W
 
 | | Structure of RyR1 (Ca2+/Caffeine/ATP/CaM1234/CHL) | | Descriptor: | 5-bromanyl-N-[4-chloranyl-2-methyl-6-(methylcarbamoyl)phenyl]-2-(3-chloranylpyridin-2-yl)pyrazole-3-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, ... | | Authors: | Ma, R, Haji-Ghassemi, O, Ma, D, Lin, L, Samurkas, A, Van Petegem, F, Yuchi, Z. | | Deposit date: | 2020-03-01 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for diamide modulation of ryanodine receptor.
Nat.Chem.Biol., 16, 2020
|
|
8HF8
 
 | | Human PPAR delta ligand binding domain in complex with a synthetic agonist V1 | | Descriptor: | 2-[4-[[2,5-bis(oxidanylidene)-3-[4-(trifluoromethyl)phenyl]imidazolidin-1-yl]methyl]-2,6-dimethyl-phenoxy]-2-methyl-propanoic acid, Peroxisome proliferator-activated receptor delta, octyl beta-D-glucopyranoside | | Authors: | Dai, L, Sun, H.B, Yuan, H.L, Feng, Z.Q. | | Deposit date: | 2022-11-09 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of the First Subnanomolar PPAR alpha / delta Dual Agonist for the Treatment of Cholestatic Liver Diseases.
J.Med.Chem., 66, 2023
|
|
7EMN
 
 | | The atomic structure of SHP2 E76A mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Luo, F, Xie, J.J, Zhu, J.D, Liu, C. | | Deposit date: | 2021-04-14 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A novel partially open state of SHP2 points to a "multiple gear" regulation mechanism.
J.Biol.Chem., 296, 2021
|
|
7X1R
 
 | | Cryo-EM structure of human thioredoxin reductase bound by Au | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GOLD ION, Thioredoxin reductase 1, ... | | Authors: | He, Z.S, Cao, P, Cao, S.H, He, B, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2022-02-24 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Au4 cluster inhibits human thioredoxin reductase activity via specifically binding of Au to Cys189
Nano Today, 47, 2022
|
|
7YQE
 
 | |
6M39
 
 | | Cryo-EM structure of SADS-CoV spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ouyang, S, Hongxin, G. | | Deposit date: | 2020-03-03 | | Release date: | 2020-08-26 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Cryo-electron Microscopy Structure of the Swine Acute Diarrhea Syndrome Coronavirus Spike Glycoprotein Provides Insights into Evolution of Unique Coronavirus Spike Proteins.
J.Virol., 94, 2020
|
|
