6CEX
 
 | |
6CF5
 
 | |
1INW
 
 | | A SIALIC ACID DERIVED PHOSPHONATE ANALOG INHIBITS DIFFERENT STRAINS OF INFLUENZA VIRUS NEURAMINIDASE WITH DIFFERENT EFFICIENCIES | | Descriptor: | (1S)-4-acetamido-1,5-anhydro-2,4-dideoxy-1-phosphono-D-glycero-D-galacto-octitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | White, C.L, Janakiraman, M.N, Laver, W.G, Philippon, C, Vasella, A, Air, G.M, Luo, M. | | Deposit date: | 1994-09-26 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A sialic acid-derived phosphonate analog inhibits different strains of influenza virus neuraminidase with different efficiencies.
J.Mol.Biol., 245, 1995
|
|
1INY
 
 | | A SIALIC ACID DERIVED PHOSPHONATE ANALOG INHIBITS DIFFERENT STRAINS OF INFLUENZA VIRUS NEURAMINIDASE WITH DIFFERENT EFFICIENCIES | | Descriptor: | (1R)-4-acetamido-1,5-anhydro-2,4-dideoxy-1-phosphono-D-glycero-D-galacto-octitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | White, C.L, Janakiraman, M.N, Laver, W.G, Philippon, C, Vasella, A, Air, G.M, Luo, M. | | Deposit date: | 1994-09-26 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A sialic acid-derived phosphonate analog inhibits different strains of influenza virus neuraminidase with different efficiencies.
J.Mol.Biol., 245, 1995
|
|
1INX
 
 | | A SIALIC ACID DERIVED PHOSPHONATE ANALOG INHIBITS DIFFERENT STRAINS OF INFLUENZA VIRUS NEURAMINIDASE WITH DIFFERENT EFFICIENCIES | | Descriptor: | (1R)-4-acetamido-1,5-anhydro-2,4-dideoxy-1-phosphono-D-glycero-D-galacto-octitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | White, C.L, Janakiraman, M.N, Laver, W.G, Philippon, C, Vasella, A, Air, G.M, Luo, M. | | Deposit date: | 1994-09-26 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A sialic acid-derived phosphonate analog inhibits different strains of influenza virus neuraminidase with different efficiencies.
J.Mol.Biol., 245, 1995
|
|
2VSU
 
 | | A ternary complex of Hydroxycinnamoyl-CoA Hydratase-Lyase (HCHL) with acetyl-Coenzyme A and vanillin gives insights into substrate specificity and mechanism. | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, ACETYL COENZYME *A, P-HYDROXYCINNAMOYL COA HYDRATASE/LYASE | | Authors: | Bennett, J.P, Bertin, L.M, Brzozowski, A.M, Walton, N.J, Grogan, G. | | Deposit date: | 2008-04-29 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Ternary Complex of Hydroxycinnamoyl-Coa Hydratase-Lyase (Hchl) with Acetyl-Coa and Vanillin Gives Insights Into Substrate Specificity and Mechanism.
Biochem.J., 414, 2008
|
|
2DND
 
 | | A BIFURCATED HYDROGEN-BONDED CONFORMATION IN THE D(A.T) BASE PAIRS OF THE DNA DODECAMER D(CGCAAATTTGCG) AND ITS COMPLEX WITH DISTAMYCIN | | Descriptor: | DISTAMYCIN A, DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Coll, M, Frederick, C.A, Wang, A.H.-J, Rich, A. | | Deposit date: | 1988-08-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin.
Proc.Natl.Acad.Sci.USA, 84, 1987
|
|
2SCU
 
 | | A detailed description of the structure of Succinyl-COA synthetase from Escherichia coli | | Descriptor: | COENZYME A, PROTEIN (SUCCINYL-COA LIGASE), SULFATE ION | | Authors: | Fraser, M.E, Wolodko, W.T, James, M.N.G, Bridger, W.A. | | Deposit date: | 1998-09-24 | | Release date: | 1999-08-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A detailed structural description of Escherichia coli succinyl-CoA synthetase.
J.Mol.Biol., 285, 1999
|
|
1BBB
 
 | |
6O9T
 
 | | KirBac3.1 mutant at a resolution of 4.1 Angstroms | | Descriptor: | 2,3,5,6-tetramethyl-1H,7H-pyrazolo[1,2-a]pyrazole-1,7-dione, Inward rectifier potassium channel Kirbac3.1, POTASSIUM ION | | Authors: | Gulbis, J.M, Black, K.A, Miller, D.M. | | Deposit date: | 2019-03-15 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.01 Å) | | Cite: | A constricted opening in Kir channels does not impede potassium conduction.
Nat Commun, 11, 2020
|
|
5KTJ
 
 | | Crystal structure of Pistol, a class of self-cleaving ribozyme | | Descriptor: | COBALT HEXAMMINE(III), MAGNESIUM ION, Pistol (50-MER), ... | | Authors: | Nguyen, L.A, Wang, J, Steitz, T.A. | | Deposit date: | 2016-07-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structure of Pistol, a class of self-cleaving ribozyme.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6RV3
 
 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation with a bound inhibitor BAY 1000493 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
6RV4
 
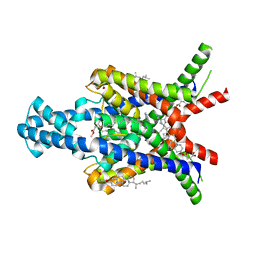 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation with a bound inhibitor BAY 2341237 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, POTASSIUM ION, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
5JTE
 
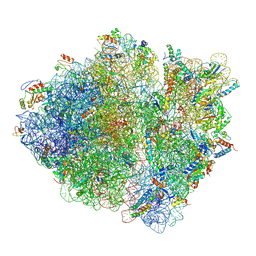 | | Cryo-EM structure of an ErmBL-stalled ribosome in complex with A-, P-, and E-tRNA | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Arenz, S, Bock, L.V, Graf, M, Innis, C.A, Beckmann, R, Grubmueller, H, Vaiana, A.C, Wilson, D.N. | | Deposit date: | 2016-05-09 | | Release date: | 2016-07-20 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A combined cryo-EM and molecular dynamics approach reveals the mechanism of ErmBL-mediated translation arrest.
Nat Commun, 7, 2016
|
|
6DTI
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with an unmodifed anticodon stem loop (ASL) of Escherichia coli transfer RNA Arginine 2 (TRNAARG2) bound to an mRNA with an CGU-codon in the A-site and paromomycin | | Descriptor: | 16s rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Cantara, W.A, DeMirci, H, Agris, P.F. | | Deposit date: | 2018-06-16 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | A Structural Basis for Restricted Codon Recognition Mediated by 2-thiocytidine in tRNA Containing a Wobble Position Inosine.
J.Mol.Biol., 432, 2020
|
|
5NP8
 
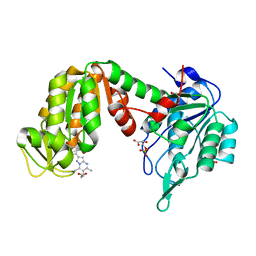 | | PGK1 in complex with CRT0063465 (3-[2-(4-bromophenyl)-5,7-dimethyl-pyrazolo[1,5-a]pyrimidin-6-yl]propanoic acid) | | Descriptor: | 1,2-ETHANEDIOL, 3-PHOSPHOGLYCERIC ACID, 3-[2-(4-bromophenyl)-5,7-dimethyl-pyrazolo[1,5-a]pyrimidin-6-yl]propanoic acid, ... | | Authors: | Turnbull, A.P, Bilsland, A.E, Liu, Y, Sumpton, D, Stevenson, K, Cairney, C.J, Roffey, J, Jenkinson, D, Keith, W.N. | | Deposit date: | 2017-04-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Pyrazolopyrimidine Ligand of Human PGK1 and Stress Sensor DJ1 Modulates the Shelterin Complex and Telomere Length Regulation.
Neoplasia, 21, 2019
|
|
6MPI
 
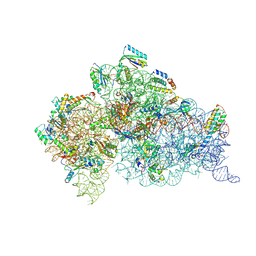 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a 2-thiocytidine (s2C32) and inosine (I34) modified anticodon stem loop (ASL) of Escherichia coli transfer RNA Arginine 1 (TRNAARG1) bound to an mRNA with an CGU-codon in the A-site and paromomycin | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Cantara, W.A, DeMirci, H, Agris, P.F. | | Deposit date: | 2018-10-07 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | A Structural Basis for Restricted Codon Recognition Mediated by 2-thiocytidine in tRNA Containing a Wobble Position Inosine.
J.Mol.Biol., 432, 2020
|
|
6MKN
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with an inosine (I34) modified anticodon stem loop (ASL) of Escherichia coli transfer RNA Arginine 2 (TRNAARG2) bound to an mRNA with an CGU-codon in the A-site and paromomycin | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Cantara, W.A, DeMirci, H, Agris, P.F. | | Deposit date: | 2018-09-25 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | A Structural Basis for Restricted Codon Recognition Mediated by 2-thiocytidine in tRNA Containing a Wobble Position Inosine.
J.Mol.Biol., 432, 2020
|
|
6MPF
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a 2-thiocytidine (s2C32) and inosine (I34) modified anticodon stem loop (ASL) of Escherichia coli transfer RNA Arginine 1 (TRNAARG1) bound to an mRNA with an CGC-codon in the A-site and paromomycin | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Cantara, W.A, DeMirci, H, Agris, P.F. | | Deposit date: | 2018-10-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | A Structural Basis for Restricted Codon Recognition Mediated by 2-thiocytidine in tRNA Containing a Wobble Position Inosine.
J.Mol.Biol., 432, 2020
|
|
5SFH
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with N-cyclopropyl-5-[2-(5,8-dimethyl-[1,2,4]triazolo[1,5-a]pyrazin-2-yl)ethyl]-N,1-dimethyl-1,2,4-triazol-3-amine | | Descriptor: | MAGNESIUM ION, N-cyclopropyl-5-{2-[(4S)-5,8-dimethyl[1,2,4]triazolo[1,5-a]pyrazin-2-yl]ethyl}-N,1-dimethyl-1H-1,2,4-triazol-3-amine, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFS
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 5,8-dimethyl-2-[2-[2-methyl-5-[(3S)-3-methylpyrrolidin-1-yl]-1,2,4-triazol-3-yl]ethyl]-[1,2,4]triazolo[1,5-a]pyrazine | | Descriptor: | (4S)-5,8-dimethyl-2-(2-{1-methyl-3-[(3S)-3-methylpyrrolidin-1-yl]-1H-1,2,4-triazol-5-yl}ethyl)[1,2,4]triazolo[1,5-a]pyrazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFT
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 5,8-dimethyl-2-[2-(1-methyl-5-pyrrolidin-1-yl-1,2,4-triazol-3-yl)ethyl]-[1,2,4]triazolo[1,5-a]pyrazine | | Descriptor: | (4S)-5,8-dimethyl-2-{2-[1-methyl-5-(pyrrolidin-1-yl)-1H-1,2,4-triazol-3-yl]ethyl}[1,2,4]triazolo[1,5-a]pyrazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
6EDD
 
 | | Crystal structure of a GNAT Superfamily PA3944 acetyltransferase in complex with CoA (P1 space group) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acetyltransferase PA3944, ... | | Authors: | Czub, M.P, Porebski, P.J, Majorek, K.A, Satchell, K.J, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-08-09 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Gcn5-Related N-Acetyltransferase (GNAT) Capable of Acetylating Polymyxin B and Colistin Antibiotics in Vitro.
Biochemistry, 57, 2018
|
|
6EDV
 
 | | Structure of a GNAT superfamily acetyltransferase PA3944 in complex with CoA | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase PA3944, CALCIUM ION, ... | | Authors: | Majorek, K.A, Satchell, K.J.F, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-08-12 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A Gcn5-Related N-Acetyltransferase (GNAT) Capable of Acetylating Polymyxin B and Colistin Antibiotics in Vitro.
Biochemistry, 57, 2018
|
|
5FGN
 
 | |
