6OQ6
 
 | |
7UBX
 
 | |
8P51
 
 | |
8P52
 
 | |
4R04
 
 | |
7POG
 
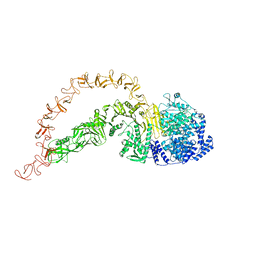 | | High-resolution structure of native toxin A from Clostridioides difficile | | Descriptor: | Toxin A, ZINC ION | | Authors: | Boesen, T, Joergensen, R, Aminzadeh, A, Engelbrecht Larsen, C. | | Deposit date: | 2021-09-08 | | Release date: | 2021-12-08 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | High-resolution structure of native toxin A from Clostridioides difficile.
Embo Rep., 23, 2022
|
|
6OQ5
 
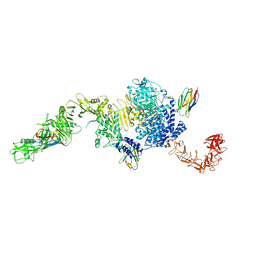 | | Structure of the full-length Clostridium difficile toxin B in complex with 3 VHHs | | Descriptor: | 5D, 7F, E3, ... | | Authors: | Chen, P, Lam, K, Jin, R. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.87 Å) | | Cite: | Structure of the full-length Clostridium difficile toxin B.
Nat.Struct.Mol.Biol., 26, 2019
|
|
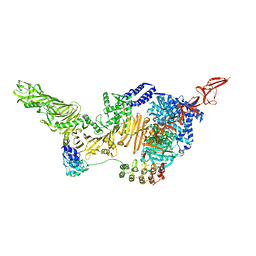
6AR6
 
 | |
6C0B
 
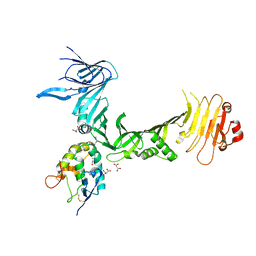 | | Structural basis for recognition of frizzled proteins by Clostridium difficile toxin B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-2, MALONATE ION, ... | | Authors: | Chen, P, Lam, K, Jin, R. | | Deposit date: | 2017-12-28 | | Release date: | 2018-05-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for recognition of frizzled proteins byClostridium difficiletoxin B.
Science, 360, 2018
|
|
7U1Z
 
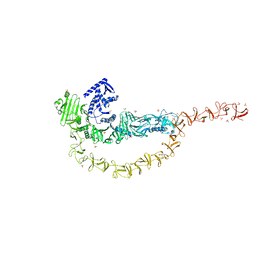 | | Crystal structure of the DRBD and CROPs of TcdA | | Descriptor: | SULFATE ION, Toxin A | | Authors: | Baohua, C, Peng, C, Kay, P, Rongsheng, J. | | Deposit date: | 2022-02-22 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure and conformational dynamics of Clostridioides difficile toxin A.
Life Sci Alliance, 5, 2022
|
|
7V1N
 
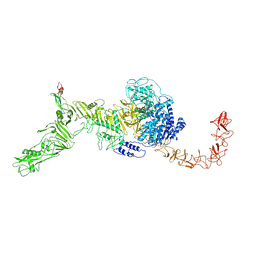 | | Structure of the Clade 2 C. difficile TcdB in complex with its receptor TFPI | | Descriptor: | Isoform Beta of Tissue factor pathway inhibitor, Toxin B | | Authors: | Luo, J, Yang, Q, Zhang, X, Zhang, Y, Wan, L, Li, Y, Tao, L. | | Deposit date: | 2021-08-05 | | Release date: | 2022-02-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | TFPI is a colonic crypt receptor for TcdB from hypervirulent clade 2 C. difficile.
Cell, 185, 2022
|
|
8QEN
 
 | |
8QEO
 
 | |
8X2I
 
 | |
8X2H
 
 | |
8JB5
 
 | |
8JHZ
 
 | | Cryo-EM structure of the TcsH-TMPRSS2 complex | | Descriptor: | Hemorrhagic toxin, Transmembrane protease serine 2, ZINC ION | | Authors: | Zhou, R, Liang, T, Zhan, X. | | Deposit date: | 2023-05-25 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of TMPRSS2 recognition by Paeniclostridium sordellii hemorrhagic toxin.
Nat Commun, 15, 2024
|
|
8P50
 
 | | Photorhabdus luminescens Makes caterpillars floppy (Mcf) toxin with the C-terminal deletion in complex with Arf3 | | Descriptor: | ADP-ribosylation factor 3, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Belyy, A, Heilen, P, Hofnagel, O, Raunser, S. | | Deposit date: | 2023-05-23 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Structure and activation mechanism of the Makes caterpillars floppy 1 toxin.
Nat Commun, 14, 2023
|
|
7ML7
 
 | |
7N8X
 
 | | Partial C. difficile TcdB and CSPG4 fragment | | Descriptor: | Chondroitin sulfate proteoglycan 4, Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-16 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
7N9Q
 
 | | State 3 of TcdB and FZD2 at pH5 | | Descriptor: | Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-18 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
7N9R
 
 | | state 4 of TcdB and FZD2 at pH5 | | Descriptor: | Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-18 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
7N95
 
 | | state 1 of TcdB and FZD2 at pH5 | | Descriptor: | Frizzled-2, Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-16 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
7N97
 
 | | State 2 of TcdB and FZD2 at pH5 | | Descriptor: | Frizzled-2, Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-17 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural Basis for Receptor Recognition of the Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
7N9Y
 
 | | Full-length TcdB and CSPG4 (401-560) complex | | Descriptor: | Chondroitin sulfate proteoglycan 4, Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-18 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
