6SWI
 
 | |
3OOU
 
 | | The structure of a protein with unkown function from Listeria innocua | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Lin2118 protein | | Authors: | Fan, Y, Mack, J, Feldman, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The structure of a protein with unkown function from Listeria innocua
To be Published
|
|
4MLO
 
 | | 1.65A resolution structure of ToxT from Vibrio cholerae (P21 Form) | | Descriptor: | CHLORIDE ION, PALMITOLEIC ACID, TCP pilus virulence regulatory protein | | Authors: | Lovell, S, Wehmeyer, G, Battaile, K.P, Li, J, Egan, S. | | Deposit date: | 2013-09-06 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 1.65 angstrom resolution structure of the AraC-family transcriptional activator ToxT from Vibrio cholerae.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
3OIO
 
 | | Crystal structure of transcriptional regulator (AraC-type DNA-binding domain-containing proteins) from Chromobacterium violaceum | | Descriptor: | CHLORIDE ION, SULFATE ION, Transcriptional regulator (AraC-type DNA-binding domain-containing proteins) | | Authors: | Chang, C, Mack, J, Feldman, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-19 | | Release date: | 2010-09-08 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of transcriptional regulator (AraC-type DNA-binding domain-containing proteins) from Chromobacterium violaceum
To be Published
|
|
6P7R
 
 | |
3MN2
 
 | |
5CHH
 
 | | Crystal structure of transcriptional regulator CdpR from Pseudomonas aeruginosa | | Descriptor: | AraC family transcriptional regulator | | Authors: | Zhao, J.R, Yu, X, Zhu, M, Kang, H.P, Kong, W.N, Ma, J.B, Deng, X, Gan, J.H, Liang, H.H. | | Deposit date: | 2015-07-10 | | Release date: | 2016-05-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Molecular Mechanism of CdpR Involved in Quorum-Sensing and Bacterial Virulence in Pseudomonas aeruginosa
Plos Biol., 14, 2016
|
|
3GBG
 
 | | Crystal Structure of ToxT from Vibrio Cholerae O395 | | Descriptor: | PALMITOLEIC ACID, TCP pilus virulence regulatory protein | | Authors: | Lowden, M.J, Kull, F.J. | | Deposit date: | 2009-02-19 | | Release date: | 2010-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5SUX
 
 | |
3LSG
 
 | |
6PB9
 
 | |
1BL0
 
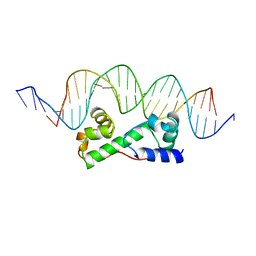 | | MULTIPLE ANTIBIOTIC RESISTANCE PROTEIN (MARA)/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*TP*GP*CP*CP*AP*CP*GP*TP*TP*TP*TP*GP*CP*TP*AP*AP*AP*TP* CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*AP*TP*TP*TP*AP*GP*CP*AP*AP*AP*AP*CP*GP*TP*GP*GP*CP*AP* TP*C)-3'), PROTEIN (MULTIPLE ANTIBIOTIC RESISTANCE PROTEIN) | | Authors: | Davies, S, Rhee, R.G, Martin, J.L, Rosner, D.R. | | Deposit date: | 1998-07-22 | | Release date: | 1998-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
5SUW
 
 | | Crystal Structure of ToxT from Vibrio Cholerae O395 bound to 3-(8-Methyl-1,2,3,4-tetrahydronaphthalen-1-yl)propanoic acid | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[(1R)-8-methyl-1,2,3,4-tetrahydronaphthalen-1-yl]propanoic acid, TCP pilus virulence regulatory protein | | Authors: | Kull, F.J, Kelley, A.R. | | Deposit date: | 2016-08-04 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of ToxT from Vibrio Cholerae O395 bound to 3-(8-Methyl-1,2,3,4-tetrahydronaphthalen-1-yl)propanoic acid
To Be Published
|
|
6P7T
 
 | |
1D5Y
 
 | | CRYSTAL STRUCTURE OF THE E. COLI ROB TRANSCRIPTION FACTOR IN COMPLEX WITH DNA | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*TP*GP*AP*CP*AP*TP*TP*CP*AP*GP*TP*GP*CP*TP*GP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*CP*AP*GP*CP*AP*CP*TP*GP*AP*AP*TP*GP*TP*CP*AP*AP*AP*G)-3'), ROB TRANSCRIPTION FACTOR | | Authors: | Kwon, H.J, Bennik, M.H.J, Demple, B, Ellenberger, T. | | Deposit date: | 1999-10-12 | | Release date: | 2000-04-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA.
Nat.Struct.Biol., 7, 2000
|
|
5NLA
 
 | |
6XIV
 
 | | SeMet-Rns, in complex with potential inhibitor | | Descriptor: | (2S)-2-hydroxybutanedioic acid, DECANOIC ACID, Regulatory protein Rns | | Authors: | Midgett, C.R, Kull, F.J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-30 | | Last modified: | 2022-02-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the master regulator Rns reveals an inhibitor of enterotoxigenic Escherichia coli virulence regulons.
Sci Rep, 11, 2021
|
|
3W6V
 
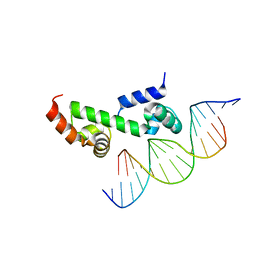 | | Crystal structure of the DNA-binding domain of AdpA, the global transcriptional factor, in complex with a target DNA | | Descriptor: | AdpA, DNA (5'-D(*AP*GP*GP*TP*TP*GP*GP*CP*GP*GP*GP*TP*TP*CP*AP*C)-3'), DNA (5'-D(*CP*TP*GP*TP*GP*AP*AP*CP*CP*CP*GP*CP*CP*AP*AP*C)-3') | | Authors: | Yao, M.D, Ohtsuka, J, Nagata, K, Miyazono, K, Ohnishi, Y, Tanokura, M. | | Deposit date: | 2013-02-22 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Complex Structure of the DNA-binding Domain of AdpA, the Global Transcription Factor in Streptomyces griseus, and a Target Duplex DNA Reveals the Structural Basis of Its Tolerant DNA Sequence Specificity
J.Biol.Chem., 288, 2013
|
|
6XIU
 
 | |
7R3W
 
 | |
7VWZ
 
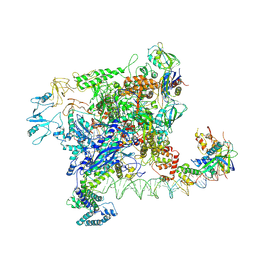 | | Cryo-EM structure of Rob-dependent transcription activation complex in a unique conformation | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Feng, Y, Shi, J. | | Deposit date: | 2021-11-12 | | Release date: | 2022-06-08 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of transcription activation by Rob, a pleiotropic AraC/XylS family regulator.
Nucleic Acids Res., 50, 2022
|
|
7BEG
 
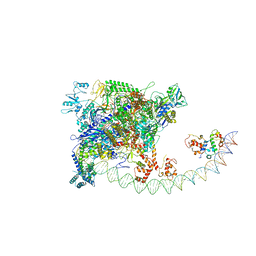 | | Structures of class I bacterial transcription complexes | | Descriptor: | Class I pacrA promoter non-template DNA, Class I pacrA promoter template DNA, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Ye, F.Z, Hao, M, Zhang, X.D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structures of Class I and Class II Transcription Complexes Reveal the Molecular Basis of RamA-Dependent Transcription Activation.
Adv Sci, 9, 2022
|
|
7BEF
 
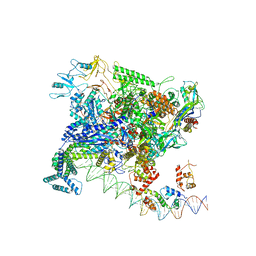 | | Structures of class II bacterial transcription complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Hao, M, Ye, F.Z, Zhang, X.D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of Class I and Class II Transcription Complexes Reveal the Molecular Basis of RamA-Dependent Transcription Activation.
Adv Sci, 9, 2022
|
|
7VWY
 
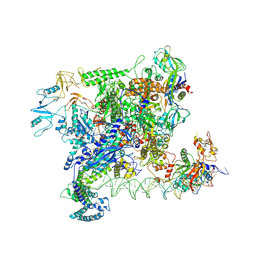 | |
1XS9
 
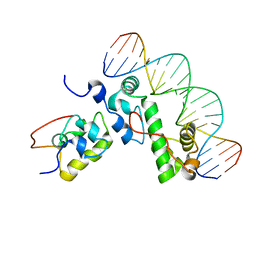 | | A MODEL OF THE TERNARY COMPLEX FORMED BETWEEN MARA, THE ALPHA-CTD OF RNA POLYMERASE AND DNA | | Descriptor: | 5'-D(P*AP*TP*GP*CP*CP*AP*CP*GP*TP*TP*TP*TP*GP*CP*TP*AP*AP*AP*TP*C)-3', 5'-D(P*GP*AP*TP*TP*TP*AP*GP*CP*AP*AP*AP*AP*CP*GP*TP*GP*GP*CP*AP*T)-3', DNA-directed RNA polymerase alpha chain, ... | | Authors: | Dangi, B, Gronenborn, A.M, Rosner, J.L, Martin, R.G. | | Deposit date: | 2004-10-18 | | Release date: | 2004-10-26 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Versatility of the carboxy-terminal domain of the alpha subunit of RNA polymerase in transcriptional activation: use of the DNA contact site as a protein contact site for MarA.
Mol.Microbiol., 54, 2004
|
|
