5UVM
 
 | | HIT family hydrolase from Clostridium thermocellum Cth-393 | | Descriptor: | ADENOSINE, Histidine triad (HIT) protein, UNKNOWN ATOM OR ION, ... | | Authors: | Habel, J, Tempel, W, Liu, Z.-J, Rose, J, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2017-02-20 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | HIT family hydrolase from Clostridium thermocellum Cth-393
To Be Published
|
|
6D6J
 
 | |
4NDI
 
 | | Human Aprataxin (Aptx) AOA1 variant K197Q bound to RNA-DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', ADENOSINE MONOPHOSPHATE, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDH
 
 | | Human Aprataxin (Aptx) bound to DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(P*GP*TP*TP*CP*TP*AP*GP*AP*AP*C)-3', ADENOSINE MONOPHOSPHATE, Aprataxin, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDG
 
 | | Human Aprataxin (Aptx) bound to RNA-DNA and Zn - adenosine vanadate transition state mimic complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', Aprataxin, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDF
 
 | | Human Aprataxin (Aptx) bound to RNA-DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', ADENOSINE MONOPHOSPHATE, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
6CVT
 
 | | Human Aprataxin (Aptx) V263G bound to RNA-DNA, AMP and Zn product complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin, DNA (5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3'), ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.941 Å) | | Cite: | Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
EMBO J., 37, 2018
|
|
6CVP
 
 | | Human Aprataxin (Aptx) R199H bound to RNA-DNA, AMP and Zn product complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin, DNA (5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3'), ... | | Authors: | Schellenberg, M.J, Williams, R.S, Tumbale, P.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
EMBO J., 37, 2018
|
|
6CVS
 
 | | Human Aprataxin (Aptx) L248M bound to DNA, AMP and Zn product | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin, BETA-MERCAPTOETHANOL, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
EMBO J., 37, 2018
|
|
6CVQ
 
 | | Human Aprataxin (Aptx) H201Q bound to RNA-DNA, AMP and Zn product complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin, BETA-MERCAPTOETHANOL, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
EMBO J., 37, 2018
|
|
6CVR
 
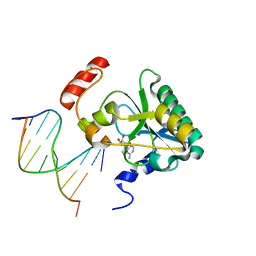 | | Human Aprataxin (Aptx) S242N bound to RNA-DNA, AMP and Zn product complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin, DNA (5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3'), ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
EMBO J., 37, 2018
|
|
6CVO
 
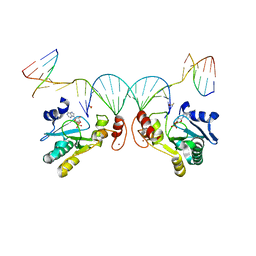 | | Human Aprataxin (Aptx) bound to nicked RNA-DNA, AMP and Zn product complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin, BETA-MERCAPTOETHANOL, ... | | Authors: | Schellenberg, M.J, Tumbale, P.P, Williams, R.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
EMBO J., 37, 2018
|
|
6GBS
 
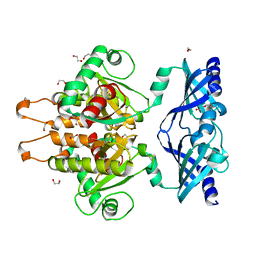 | | Crystal Structure of the C. themophilum Scavenger Decapping Enzyme DcpS apo form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Putative mRNA decapping protein | | Authors: | Fuchs, A.-L, Neu, A, Sprangers, R. | | Deposit date: | 2018-04-16 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | Molecular basis of the selective processing of short mRNA substrates by the DcpS mRNA decapping enzyme.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5BV3
 
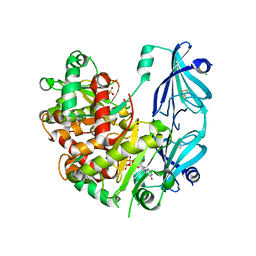 | | Yeast Scavenger Decapping Enzyme in complex with m7GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, CHLORIDE ION, SULFATE ION, ... | | Authors: | Neu, A, Neu, U, Sprangers, R. | | Deposit date: | 2015-06-04 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | An excess of catalytically required motions inhibits the scavenger decapping enzyme.
Nat.Chem.Biol., 11, 2015
|
|
6TRQ
 
 | | S.c. Scavenger Decapping Enzyme DcpS in complex with the capped RNA dinucleotide m7G-GU | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, L-GUANOSINE-5'-MONOPHOSPHATE, PHOSPHONATE, ... | | Authors: | Fuchs, A.-L, Neu, A, Sprangers, R. | | Deposit date: | 2019-12-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.944 Å) | | Cite: | Molecular basis of the selective processing of short mRNA substrates by the DcpS mRNA decapping enzyme.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5OSY
 
 | | Human Decapping Scavenger enzyme (hDcpS) in complex with m7G(5'S)ppSp(5'S)G mRNA 5' cap analog | | Descriptor: | GLYCEROL, [(2~{S},3~{S},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl-[[[(2~{S},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl-oxidanyl-phosphoryl]oxy-sulfanyl-phosphoryl]oxy-phosphinic acid, m7GpppX diphosphatase | | Authors: | Warminski, M, Nowak, E, Wojtczak, B.A, Fac-Dabrowska, K, Kubacka, D, Nowicka, A, Sikorski, P.J, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2017-08-18 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | 5'-Phosphorothiolate Dinucleotide Cap Analogues: Reagents for Messenger RNA Modification and Potent Small-Molecular Inhibitors of Decapping Enzymes.
J. Am. Chem. Soc., 140, 2018
|
|
1ST0
 
 | | Structure of DcpS bound to m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, YTTRIUM (III) ION, mRNA decapping enzyme | | Authors: | Gu, M, Fabrega, C, Liu, S.W, Liu, H, Kiledjian, M, Lima, C.D. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the structure, mechanism, and regulation of scavenger mRNA decapping activity
Mol.Cell, 14, 2004
|
|
1ST4
 
 | | Structure of DcpS bound to m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, YTTRIUM (III) ION, mRNA decapping enzyme | | Authors: | Gu, M, Fabrega, C, Liu, S.W, Liu, H, Kiledjian, M, Lima, C.D. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Insights into the structure, mechanism, and regulation of scavenger mRNA decapping activity
Mol.Cell, 14, 2004
|
|
3BLA
 
 | | Synthetic Gene Encoded DcpS bound to inhibitor DG153249 | | Descriptor: | 5-[(1S)-1-(3-chlorophenyl)ethoxy]quinazoline-2,4-diamine, Scavenger mRNA-decapping enzyme DcpS | | Authors: | Staker, B.L, Christensen, J, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-12-10 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DcpS as a therapeutic target for spinal muscular atrophy.
Acs Chem.Biol., 3, 2008
|
|
3BL9
 
 | | Synthetic Gene Encoded DcpS bound to inhibitor DG157493 | | Descriptor: | 5-{[1-(2,3-dichlorobenzyl)piperidin-4-yl]methoxy}quinazoline-2,4-diamine, Scavenger mRNA-decapping enzyme DcpS | | Authors: | Staker, B.L, Christensen, J, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-12-10 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DcpS as a therapeutic target for spinal muscular atrophy.
Acs Chem.Biol., 3, 2008
|
|
3BL7
 
 | | Synthetic Gene Encoded DcpS bound to inhibitor DG156844 | | Descriptor: | 5-{[1-(2-fluorobenzyl)piperidin-4-yl]methoxy}quinazoline-2,4-diamine, Scavenger mRNA-decapping enzyme DcpS | | Authors: | Staker, B.L, Christensen, J, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-12-10 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | DcpS as a therapeutic target for spinal muscular atrophy.
Acs Chem.Biol., 3, 2008
|
|
1VLR
 
 | |
1XMM
 
 | | Structure of human Dcps bound to m7GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, N7-METHYL-GUANOSINE-5'-MONOPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Chen, N, Song, H. | | Deposit date: | 2004-10-04 | | Release date: | 2005-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human DcpS in ligand-free and m7GDP-bound forms suggest a dynamic mechanism for scavenger mRNA decapping.
J.Mol.Biol., 347, 2005
|
|
1XML
 
 | | Structure of human Dcps | | Descriptor: | PHOSPHATE ION, heat shock-like protein 1 | | Authors: | Chen, N, Song, H. | | Deposit date: | 2004-10-04 | | Release date: | 2005-03-22 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human DcpS in ligand-free and m7GDP-bound forms suggest a dynamic mechanism for scavenger mRNA decapping.
J.Mol.Biol., 347, 2005
|
|
4QEB
 
 | | Dcps in complex with covalent inhibitor targeting Tyrosine | | Descriptor: | 3-{[(2,4-diaminoquinazolin-5-yl)oxy]methyl}benzenesulfonic acid, PHOSPHATE ION, m7GpppX diphosphatase | | Authors: | Liu, S. | | Deposit date: | 2014-05-15 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Human Dcps in complex with covalent inhibitor
To be Published
|
|
