1L3A
 
 | |
4KOO
 
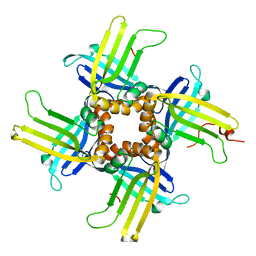
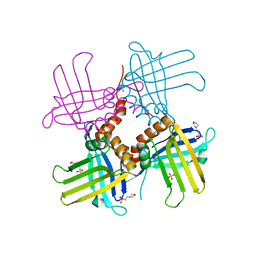 | | Crystal Structure of WHY1 from Arabidopsis thaliana | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Cappadocia, L, Parent, J.S, Brisson, N, Sygusch, J. | | Deposit date: | 2013-05-12 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A family portrait: structural comparison of the Whirly proteins from Arabidopsis thaliana and Solanum tuberosum.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4KOP
 
 | | Crystal Structure of WHY2 from Arabidopsis thaliana | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Single-stranded DNA-binding protein WHY2, mitochondrial | | Authors: | Cappadocia, L, Parent, J.S, Brisson, N, Sygusch, J. | | Deposit date: | 2013-05-12 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A family portrait: structural comparison of the Whirly proteins from Arabidopsis thaliana and Solanum tuberosum.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4KOQ
 
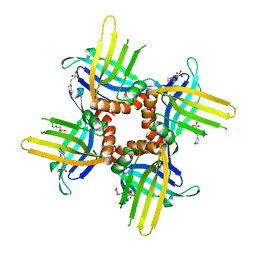
 | | Crystal Structure of WHY3 from Arabidopsis thaliana | | Descriptor: | PHOSPHATE ION, Single-stranded DNA-binding protein WHY3, chloroplastic | | Authors: | Cappadocia, L, Parent, J.S, Brisson, N, Sygusch, J. | | Deposit date: | 2013-05-12 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A family portrait: structural comparison of the Whirly proteins from Arabidopsis thaliana and Solanum tuberosum.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3N1J
 
 | |
3N1I
 
 | |
3N1L
 
 | |
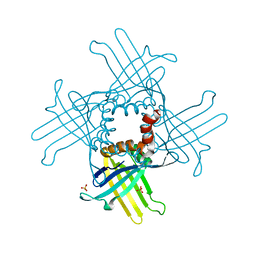
3N1H
 
 | | Crystal Structure of StWhy2 | | Descriptor: | PHOSPHATE ION, StWhy2 | | Authors: | Cappadocia, L, Brisson, N, Sygusch, J. | | Deposit date: | 2010-05-15 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Crystal Structures of DNA-Whirly Complexes and Their Role in Arabidopsis Organelle Genome Repair.
Plant Cell, 22, 2010
|
|
3N1K
 
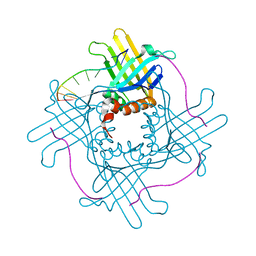 | |
3RA0
 
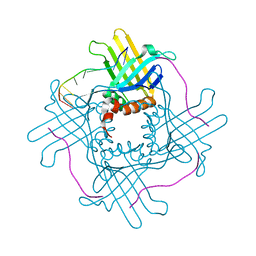 | |
3R9Y
 
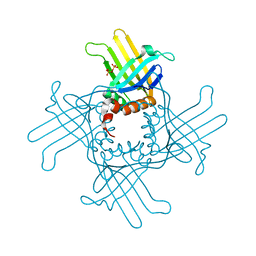 | | Crystal Structure of StWhy2 K67A (form I) | | Descriptor: | PHOSPHATE ION, Why2 protein | | Authors: | Cappadocia, L, Brisson, N, Sygusch, J. | | Deposit date: | 2011-03-26 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A conserved lysine residue of plant Whirly proteins is necessary for higher order protein assembly and protection against DNA damage.
Nucleic Acids Res., 40, 2012
|
|
3R9Z
 
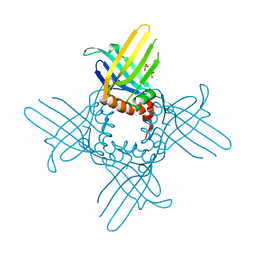 | | Crystal Structure of StWhy2 K67A (form II) | | Descriptor: | PHOSPHATE ION, Why2 protein | | Authors: | Cappadocia, L, Brisson, N, Sygusch, J. | | Deposit date: | 2011-03-26 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | A conserved lysine residue of plant Whirly proteins is necessary for higher order protein assembly and protection against DNA damage.
Nucleic Acids Res., 40, 2012
|
|
