1J8D
 
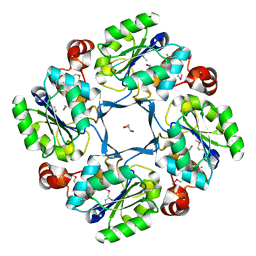 | |
1K1E
 
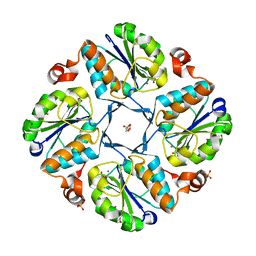 | |
1KYT
 
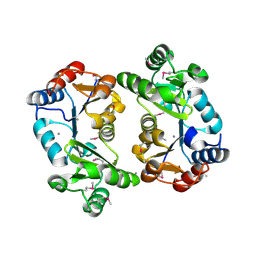 | | Crystal Structure of Thermoplasma acidophilum 0175 (APC014) | | Descriptor: | CALCIUM ION, hypothetical protein TA0175 | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Xu, X, Pennycooke, M, Gu, J, Cheung, F, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-02-05 | | Release date: | 2003-01-21 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Thermoplasma acidophilum 0175 (APC014)
To be published
|
|
1L6R
 
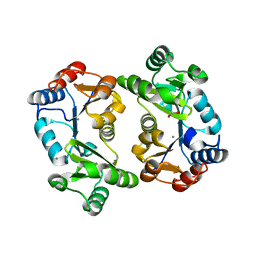 | | Crystal Structure of Thermoplasma acidophilum 0175 (APC0014) | | Descriptor: | CALCIUM ION, FORMIC ACID, HYPOTHETICAL PROTEIN TA0175 | | Authors: | Kim, Y, Joachimiak, A, Edwards, A.M, Xu, X, Pennycooke, M, Gu, J, Cheung, F, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-03-13 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure- and function-based characterization of a new phosphoglycolate phosphatase from Thermoplasma acidophilum.
J.Biol.Chem., 279, 2004
|
|
1NF2
 
 | |
1XPJ
 
 | | Crystal Structure of MCSG Target APC26283 from Vibrio cholerae | | Descriptor: | L(+)-TARTARIC ACID, MERCURY (II) ION, hypothetical protein | | Authors: | Brunzelle, J.S, Minasov, G, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-10-08 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of MCSG Target APC26283 from Vibrio cholerae
To be Published
|
|
1RLM
 
 | | Crystal Structure of ybiV from Escherichia coli K12 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphatase | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
1RLO
 
 | | Phospho-aspartyl Intermediate Analogue of ybiV from E. coli K12 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphatase | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
1RLT
 
 | | Transition State Analogue of ybiV from E. coli K12 | | Descriptor: | ACETATE ION, ALUMINUM FLUORIDE, GLYCEROL, ... | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
1XVI
 
 | | Crystal Structure of YedP, phosphatase-like domain protein from Escherichia coli K12 | | Descriptor: | PENTAETHYLENE GLYCOL, Putative mannosyl-3-phosphoglycerate phosphatase, SULFATE ION, ... | | Authors: | Kim, Y, Joachimiak, A, Cymborowski, M, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-10-28 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structure of YedP, phosphatase-like domain protein from Escherichia coli K12
To be Published
|
|
1YMQ
 
 | |
2B30
 
 | |
1WZC
 
 | | Crystal structure of Pyrococcus horikoshii mannosyl-3-phosphoglycerate phosphatase complexed with MG2+ and phosphate | | Descriptor: | MAGNESIUM ION, Mannosyl-3-phosphoglycerate phosphatase, PHOSPHATE ION | | Authors: | Kawamura, T, Watanabe, N, Tanaka, I. | | Deposit date: | 2005-03-04 | | Release date: | 2006-02-28 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of mannosyl-3-phosphoglycerate phosphatase from Pyrococcus horikoshii.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2HF2
 
 | |
2P9J
 
 | | Crystal structure of AQ2171 from Aquifex aeolicus | | Descriptor: | Hypothetical protein AQ2171 | | Authors: | Yang, H, Chen, L, Agari, Y, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2007-05-29 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of AQ2171 from Aquifex aeolicus
To be Published
|
|
2RB5
 
 | |
2RAR
 
 | |
2RAV
 
 | |
2RBK
 
 | |
2PQ0
 
 | | Crystal structure of Hyopthetical protein (gk_1056) from geobacillus Kaustophilus HTA426 | | Descriptor: | Hypothetical conserved protein GK1056 | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Kavyashree, M, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-01 | | Release date: | 2008-05-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Hyopthetical protein (gk_1056) from geobacillus Kaustophilus HTA426
To be Published
|
|
3DAO
 
 | |
3DNP
 
 | | Crystal structure of Stress response protein yhaX from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Stress response protein yhaX | | Authors: | Chang, C, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-02 | | Release date: | 2008-07-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Stress response protein yhaX from Bacillus subtilis
To be Published
|
|
2QYH
 
 | | Crystal structure of the hypothetical protein (gk1056) from geobacillus kaustophilus HTA426 | | Descriptor: | GLYCEROL, Hypothetical conserved protein, GK1056 | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-08-15 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the hypothetical protein (gk1056) from geobacillus kaustophilus HTA426
To be Published
|
|
2R8Z
 
 | | Crystal structure of YrbI phosphatase from Escherichia coli in complex with a phosphate and a calcium ion | | Descriptor: | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase, CALCIUM ION, PHOSPHATE ION | | Authors: | Tsodikov, O.V, Aggarwal, P, Rubin, J.R, Stuckey, J.A, Woodard, R.W, Biswas, T. | | Deposit date: | 2007-09-11 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Tail of KdsC: CONFORMATIONAL CHANGES CONTROL THE ACTIVITY OF A HALOACID DEHALOGENASE SUPERFAMILY PHOSPHATASE.
J.Biol.Chem., 284, 2009
|
|
2R8X
 
 | | Crystal structure of YrbI phosphatase from Escherichia coli | | Descriptor: | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase, CHLORIDE ION | | Authors: | Tsodikov, O.V, Aggarwal, P, Rubin, J.R, Stuckey, J.A, Woodard, R.W, Biswas, T. | | Deposit date: | 2007-09-11 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Tail of KdsC: CONFORMATIONAL CHANGES CONTROL THE ACTIVITY OF A HALOACID DEHALOGENASE SUPERFAMILY PHOSPHATASE.
J.Biol.Chem., 284, 2009
|
|
