1Z0W
 
 | | Crystal Structure of A. fulgidus Lon proteolytic domain at 1.2A resolution | | Descriptor: | CALCIUM ION, Putative protease La homolog type | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | Deposit date: | 2005-03-02 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
1Z0C
 
 | | Crystal Structure of A. fulgidus Lon proteolytic domain D508A mutant | | Descriptor: | Putative protease La homolog type | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | Deposit date: | 2005-03-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
1Z0B
 
 | | Crystal Structure of A. fulgidus Lon proteolytic domain E506A mutant | | Descriptor: | CALCIUM ION, Putative protease La homolog type | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | Deposit date: | 2005-03-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
1RRE
 
 | | Crystal structure of E.coli Lon proteolytic domain | | Descriptor: | ATP-dependent protease La, SULFATE ION | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Rasulova, F, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
4YPM
 
 | |
8DVH
 
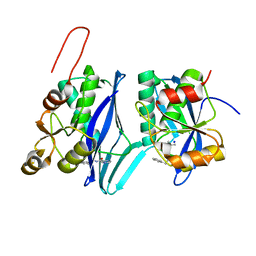 | | Crystal structure of ATP-dependent Lon protease from Bacillus subtillis (BsLonBA) | | Descriptor: | Lon protease 2, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, SODIUM ION | | Authors: | Sekula, B, Li, M, Gustchina, A, Wlodawer, A. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unique Structural Fold of LonBA Protease from Bacillus subtilis, a Member of a Newly Identified Subfamily of Lon Proteases.
Int J Mol Sci, 23, 2022
|
|
1XHK
 
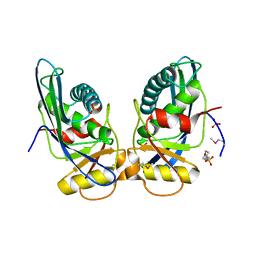 | | Crystal structure of M. jannaschii Lon proteolytic domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative protease La homolog, SULFATE ION | | Authors: | Im, Y.J, Na, Y, Kang, G.B, Rho, S.-H, Kim, M.-K, Lee, J.H, Chung, C.H, Eom, S.H. | | Deposit date: | 2004-09-20 | | Release date: | 2004-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The active site of a lon protease from Methanococcus jannaschii distinctly differs from the canonical catalytic Dyad of Lon proteases.
J.Biol.Chem., 279, 2004
|
|
4FWG
 
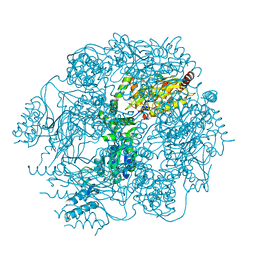 | | Crystal structure of the Lon-like protease MtaLonC in complex with lactacystin | | Descriptor: | Omuralide, open form, PHOSPHATE ION, ... | | Authors: | Chang, C.I, Kuo, C.I, Huang, K.F. | | Deposit date: | 2012-07-01 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structures of an ATP-independent Lon-like protease and its complexes with covalent inhibitors
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4FW9
 
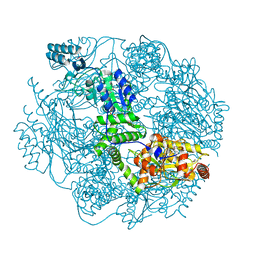 | | Crystal structure of the Lon-like protease MtaLonC | | Descriptor: | PHOSPHATE ION, TTC1975 peptidase | | Authors: | Chang, C.I, Ihara, K, Kuo, C.I, Huang, K.F, Wakatsuki, S. | | Deposit date: | 2012-06-30 | | Release date: | 2013-06-26 | | Last modified: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of an ATP-independent Lon-like protease and its complexes with covalent inhibitors
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2X36
 
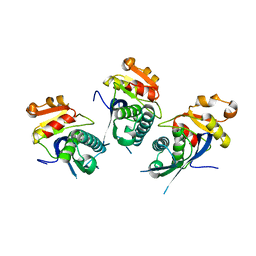 | | Structure of the proteolytic domain of the Human Mitochondrial Lon protease | | Descriptor: | LON PROTEASE HOMOLOG, MITOCHONDRIAL | | Authors: | Garcia, J, Ondrovicova, G, Blagova, E, Levdikov, V.M, Bauer, J.A, Kutejova, E, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2010-01-21 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Catalytic Domain of the Human Mitochondrial Lon Protease: Proposed Relation of Oligomer Formation and Activity.
Protein Sci., 19, 2010
|
|
3K1J
 
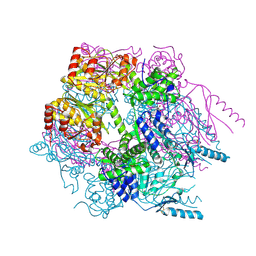 | | Crystal structure of Lon protease from Thermococcus onnurineus NA1 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Cha, S.S, An, Y.J. | | Deposit date: | 2009-09-28 | | Release date: | 2010-09-22 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Lon protease: molecular architecture of gated entry to a sequestered degradation chamber
Embo J., 29, 2010
|
|
4FWD
 
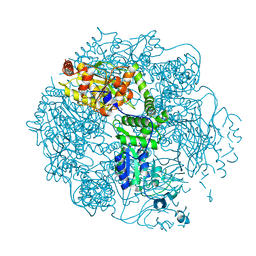 | | Crystal structure of the Lon-like protease MtaLonC in complex with bortezomib | | Descriptor: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, PHOSPHATE ION, TTC1975 peptidase | | Authors: | Chang, C.I, Kuo, C.I, Huang, K.F. | | Deposit date: | 2012-06-30 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of an ATP-independent Lon-like protease and its complexes with covalent inhibitors
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1Z0E
 
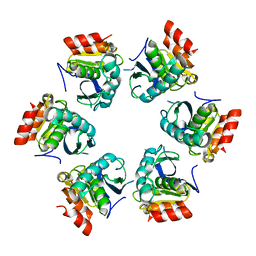 | | Crystal Structure of A. fulgidus Lon proteolytic domain | | Descriptor: | Putative protease La homolog type | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | Deposit date: | 2005-03-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
7EV6
 
 | | Crystal structure of the Lon-like protease MtaLonC with D581A mutation in complex with F-b20-Q | | Descriptor: | Endopeptidase La, F-b20-Q peptide {ortho-aminobenzoic acid (Abz)- QLRSLNGEWRFAWFPAPEAV[Tyr(3-NO2)]A}, PHOSPHATE ION | | Authors: | Hsieh, K.Y, Kuo, C.I, Su, S.C, Huang, K.F, Chang, C.I. | | Deposit date: | 2021-05-20 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
1RR9
 
 | | Catalytic domain of E.coli Lon protease | | Descriptor: | ATP-dependent protease La, SULFATE ION | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | Deposit date: | 2003-12-08 | | Release date: | 2003-12-23 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
7EV4
 
 | | Crystal structure of the Lon-like protease MtaLonC with S582A mutation in complex with F-b20-Q | | Descriptor: | Endopeptidase La, F-b20-Q peptide {ortho-aminobenzoic acid (Abz)- QLRSLNGEWRFAWFPAPEAV[Tyr(3-NO2)]A}, PHOSPHATE ION | | Authors: | Hsieh, K.Y, Kuo, C.I, Su, S.C, Huang, K.F, Chang, C.I. | | Deposit date: | 2021-05-20 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
6X27
 
 | |
4FWH
 
 | | Crystal structure of the Lon-like protease MtaLonC in complex with MG262 | | Descriptor: | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(1R)-1-(dihydroxyboranyl)-3-methylbutyl]-L-leucinamide, PHOSPHATE ION, TTC1975 peptidase | | Authors: | Chang, C.I, Kuo, C.I, Huang, K.F. | | Deposit date: | 2012-07-01 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structures of an ATP-independent Lon-like protease and its complexes with covalent inhibitors
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7EUY
 
 | | Crystal structure of the Lon-like protease MtaLonC with D582A mutation in complex with substrate polypeptide | | Descriptor: | ALA-PRO-GLU-ALA-VAL, Endopeptidase La, PHOSPHATE ION | | Authors: | Hsieh, K.Y, Kuo, C.I, Su, S.C, Huang, K.F, Chang, C.I. | | Deposit date: | 2021-05-19 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
6WYS
 
 | | Lon protease proteolytic domain | | Descriptor: | Lon protease homolog, mitochondrial, SULFATE ION | | Authors: | Lee, C.C, Spraggon, G. | | Deposit date: | 2020-05-13 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.229 Å) | | Cite: | Structure-Based Design of Selective LONP1 Inhibitors for Probing In Vitro Biology.
J.Med.Chem., 64, 2021
|
|
7EUX
 
 | | Crystal structure of the Lon-like protease MtaLonC with D581A mutation in complex with substrate polypeptide | | Descriptor: | ALA-PRO-GLU-ALA-VAL, Endopeptidase La, PHOSPHATE ION | | Authors: | Hsieh, K.Y, Kuo, C.I, Su, S.C, Huang, K.F, Chang, C.I. | | Deposit date: | 2021-05-19 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
8EIL
 
 | | C-Terminal Domain of BrxL from Acinetobacter BREX type I phage restriction system | | Descriptor: | MALONIC ACID, Protease Lon-related BREX system protein BrxL, SUCCINIC ACID | | Authors: | Doyle, L.A, Stoddard, B.L, Kaiser, B. | | Deposit date: | 2022-09-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure, substrate binding and activity of a unique AAA+ protein: the BrxL phage restriction factor.
Nucleic Acids Res., 51, 2023
|
|
1Z0G
 
 | | Crystal Structure of A. fulgidus Lon proteolytic domain | | Descriptor: | Putative protease La homolog type | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | Deposit date: | 2005-03-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
7FIE
 
 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex (conformation 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | Deposit date: | 2021-07-31 | | Release date: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7FD4
 
 | | A complete three-dimensional structure of the Lon protease translocating a protein substrate (conformation 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Lon protease, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Pintilie, G, Zhang, K, Chang, C. | | Deposit date: | 2021-07-16 | | Release date: | 2021-11-03 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Complete three-dimensional structures of the Lon protease translocating a protein substrate.
Sci Adv, 7, 2021
|
|
