4WIT
 
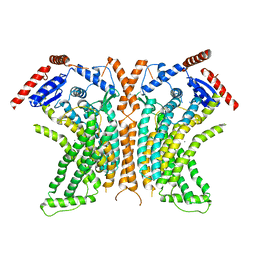 | | TMEM16 lipid scramblase in crystal form 2 | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Dutzler, R, Brunner, J.D, Lim, N.K, Schenck, S. | | Deposit date: | 2014-09-26 | | Release date: | 2014-11-12 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | X-ray structure of a calcium-activated TMEM16 lipid scramblase.
Nature, 516, 2014
|
|
4WIS
 
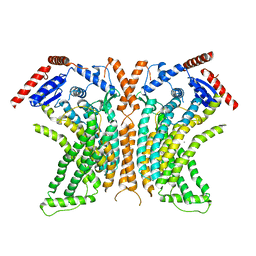 | | Crystal structure of the lipid scramblase nhTMEM16 in crystal form 1 | | Descriptor: | CALCIUM ION, lipid scramblase | | Authors: | Dutzler, R, Brunner, J.D, Lim, N.K, Schenck, S. | | Deposit date: | 2014-09-26 | | Release date: | 2014-11-12 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray structure of a calcium-activated TMEM16 lipid scramblase.
Nature, 516, 2014
|
|
5NL2
 
 | | cryo-EM structure of the mTMEM16A ion channel at 6.6 A resolution. | | Descriptor: | Anoctamin-1 | | Authors: | Paulino, C, Neldner, Y, Lam, K.M, Kalienkova, V, Brunner, J.D, Schenck, S, Dutzler, R. | | Deposit date: | 2017-04-03 | | Release date: | 2017-06-07 | | Last modified: | 2018-02-28 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural basis for anion conduction in the calcium-activated chloride channel TMEM16A.
Elife, 6, 2017
|
|
5OYB
 
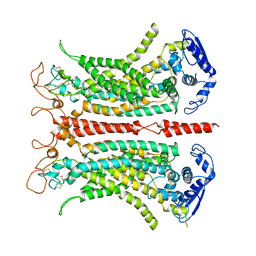 | | Structure of calcium-bound mTMEM16A chloride channel at 3.75 A resolution | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Paulino, C, Kalienkova, V, Lam, K.M, Neldner, Y, Dutzler, R. | | Deposit date: | 2017-09-08 | | Release date: | 2017-12-20 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM.
Nature, 552, 2017
|
|
5OYG
 
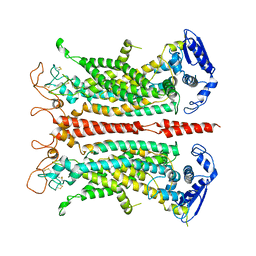 | | Structure of calcium-free mTMEM16A chloride channel at 4.06 A resolution | | Descriptor: | Anoctamin-1 | | Authors: | Paulino, C, Kalienkova, V, Lam, K.M, Neldner, Y, Dutzler, R. | | Deposit date: | 2017-09-08 | | Release date: | 2017-12-20 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM.
Nature, 552, 2017
|
|
6BGJ
 
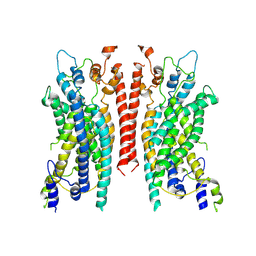 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in LMNG | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, L, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
6BGI
 
 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in nanodisc | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, J, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
5OC9
 
 | | Crystal Structure of human TMEM16K / Anoctamin 10 | | Descriptor: | (2R)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, Anoctamin-10, CALCIUM ION | | Authors: | Bushell, S.R, Pike, A.C.W, Chu, A, Tessitore, A, Rotty, B, Mukhopadhyay, S, Kupinska, K, Shrestha, L, Borkowska, O, Chalk, R, Burgess-Brown, N.A, Love, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-29 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
6E1O
 
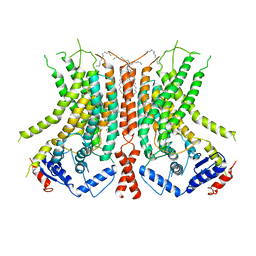 | |
6E0H
 
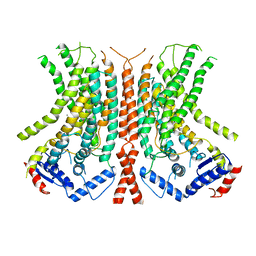 | |
6DZ7
 
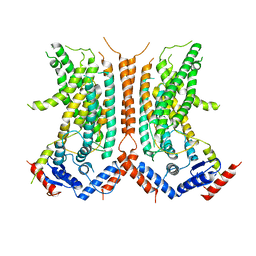 | |
6QMB
 
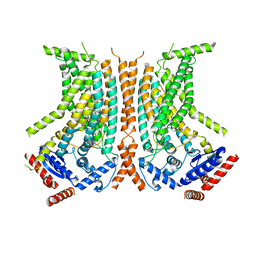 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in nanodisc (closed state) | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QPB
 
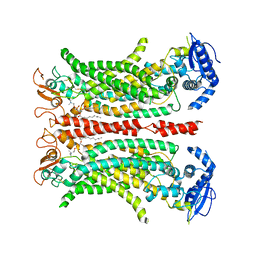 | | Cryo-EM structure of calcium-free mTMEM16F lipid scramblase in digitonin | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6 | | Authors: | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-13 | | Release date: | 2019-03-06 | | Last modified: | 2019-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
6QM4
 
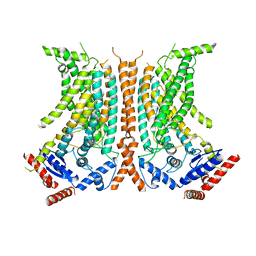 | | Cryo-EM structure of calcium-free nhTMEM16 lipid scramblase in nanodisc | | Descriptor: | Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QM9
 
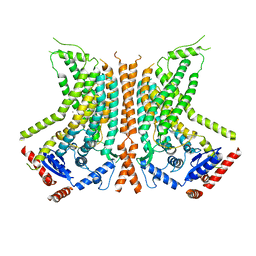 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in nanodisc (open state) | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QP6
 
 | | Cryo-EM structure of calcium-bound mTMEM16F lipid scramblase in digitonin | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6, CALCIUM ION | | Authors: | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-13 | | Release date: | 2019-03-06 | | Last modified: | 2019-04-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
6QM6
 
 | | Cryo-EM structure of calcium-free nhTMEM16 lipid scramblase in DDM | | Descriptor: | Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QMA
 
 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in nanodisc (intermediate state) | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QPC
 
 | | Cryo-EM structure of calcium-bound mTMEM16F lipid scramblase in nanodisc | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6, CALCIUM ION | | Authors: | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-13 | | Release date: | 2019-03-06 | | Last modified: | 2019-04-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
6QPI
 
 | | Cryo-EM structure of calcium-free mTMEM16F lipid scramblase in nanodisc | | Descriptor: | Anoctamin-6 | | Authors: | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-06 | | Last modified: | 2019-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
6QM5
 
 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in DDM | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6R7X
 
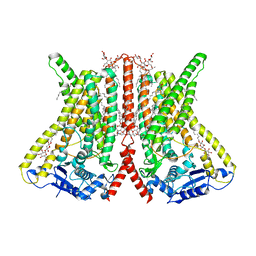 | | CryoEM structure of calcium-bound human TMEM16K / Anoctamin 10 in detergent (2mM Ca2+, closed form) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-10, CALCIUM ION, ... | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Baronina, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
6R7Y
 
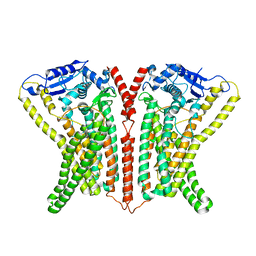 | | CryoEM structure of calcium-bound human TMEM16K / Anoctamin 10 in detergent (low Ca2+, closed form) | | Descriptor: | Anoctamin-10, CALCIUM ION | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
6R65
 
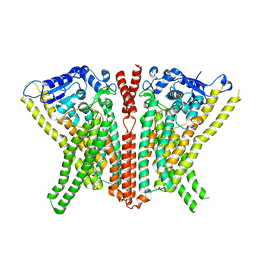 | | Crystal Structure of human TMEM16K / Anoctamin 10 (Form 2) | | Descriptor: | Anoctamin-10, CALCIUM ION | | Authors: | Bushell, S.R, Pike, A.C.W, Chu, A, Tessitore, A, Rotty, B, Mukhopadhyay, S, Kupinska, K, Shrestha, L, Borkowska, O, Chalk, R, Burgess-Brown, N.A, Love, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-26 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
6R7Z
 
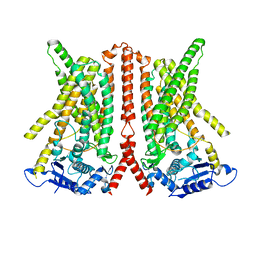 | | CryoEM structure of calcium-free human TMEM16K / Anoctamin 10 in detergent (closed form) | | Descriptor: | Anoctamin-10 | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (5.14 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
