8WXP
 
 | |
8WXO
 
 | |
8WXN
 
 | |
8WXM
 
 | |
8WGL
 
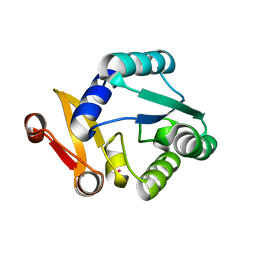 | | Crystal structure of Rhodothermus marinus substrate-binding protein (Hg soaking) | | Descriptor: | ABC-type uncharacterized transport system periplasmic component-like protein, MERCURY (II) ION | | Authors: | Nam, K.H. | | Deposit date: | 2023-09-22 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and bioinformatics analysis of single-domain substrate-binding protein from Rhodothermus marinus.
Biochem Biophys Rep, 37, 2024
|
|
8WGK
 
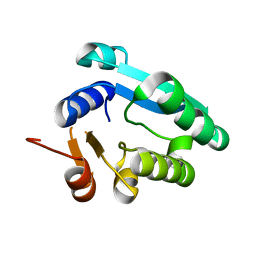 | |
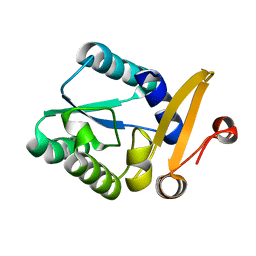
6K1Y
 
 | |
6K1X
 
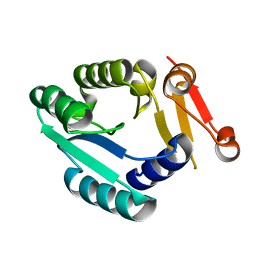 | |
6K1W
 
 | |
6HNK
 
 | | The ligand-free, open structure of CD0873, a substrate binding protein with adhesive properties from Clostridium difficile. | | Descriptor: | ABC-type transport system, sugar-family extracellular solute-binding protein | | Authors: | Bradshaw, W.J, Kovacs-Simon, A, Harmer, N.J, Michell, S.L, Acharya, K.R. | | Deposit date: | 2018-09-15 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular features of lipoprotein CD0873: A potential vaccine against the human pathogenClostridioides difficile.
J.Biol.Chem., 294, 2019
|
|
6HNJ
 
 | | The ligand-bound, open structure of CD0873, a substrate binding protein with adhesive properties from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, ABC-type transport system, sugar-family extracellular solute-binding protein, ... | | Authors: | Bradshaw, W.J, Kovacs-Simon, A, Harmer, N.J, Michell, S.L, Acharya, K.R. | | Deposit date: | 2018-09-15 | | Release date: | 2019-08-28 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular features of lipoprotein CD0873: A potential vaccine against the human pathogenClostridioides difficile.
J.Biol.Chem., 294, 2019
|
|
6HNI
 
 | | The ligand-bound, closed structure of CD0873, a substrate binding protein with adhesive properties from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, ABC-type transport system, sugar-family extracellular solute-binding protein, ... | | Authors: | Bradshaw, W.J, Kovacs-Simon, A, Harmer, N.J, Michell, S.L, Acharya, K.R. | | Deposit date: | 2018-09-15 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Molecular features of lipoprotein CD0873: A potential vaccine against the human pathogenClostridioides difficile.
J.Biol.Chem., 294, 2019
|
|
5Z6V
 
 | |
3LKV
 
 | | Crystal structure of conserved domain protein from vibrio cholerae o1 biovar eltor str. n16961 | | Descriptor: | GLYCEROL, PHENYLALANINE, SULFATE ION, ... | | Authors: | Nocek, B, Duggan, E, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-27 | | Release date: | 2010-03-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of conserved domain protein from vibrio cholerae o1 biovar eltor str. n16961
To be Published
|
|
3LFT
 
 | |
