2FGS
 
 | | Crystal structure of Campylobacter jejuni YCEI protein, structural genomics | | Descriptor: | Putative periplasmic protein, SULFATE ION | | Authors: | Patskovsky, Y, Ramagopal, U, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-12-22 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Campylobacter Jejuni YceI Periplasmic Protein
To be Published
|
|
7BWL
 
 | | Structure of antibiotic sequester from Pseudomonas aerurinosa | | Descriptor: | UPF0312 protein PA0423, Ubiquinone-8 | | Authors: | Hong, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the Pseudomonas aeruginosa PA0423 protein and its functional implication in antibiotic sequestration.
Biochem.Biophys.Res.Commun., 528, 2020
|
|
3HPE
 
 | |
5IXG
 
 | | Crystal Structure of Burkholderia cenocepacia BcnB | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, YceI | | Authors: | Loutet, S.A, Murphy, M.E.P. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Antibiotic Capture by Bacterial Lipocalins Uncovers an Extracellular Mechanism of Intrinsic Antibiotic Resistance.
MBio, 8, 2017
|
|
3Q34
 
 | |
2X34
 
 | | Structure of a polyisoprenoid binding domain from Saccharophagus degradans implicated in plant cell wall breakdown | | Descriptor: | CELLULOSE-BINDING PROTEIN, X158, Ubiquinone-8 | | Authors: | Vincent, F, Dal Molin, D, Weiner, R.M, Bourne, Y, Henrissat, B. | | Deposit date: | 2010-01-19 | | Release date: | 2010-03-23 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Polyisoprenoid Binding Domain from Saccharophagus Degradans Implicated in Plant Cell Wall Breakdown
FEBS Lett., 584, 2010
|
|
2X32
 
 | | Structure of a polyisoprenoid binding domain from Saccharophagus degradans implicated in plant cell wall breakdown | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, CELLULOSE-BINDING PROTEIN, IMIDAZOLE | | Authors: | Vincent, F, Dal Molin, D, Weiner, R.M, Bourne, Y, Henrissat, B. | | Deposit date: | 2010-01-19 | | Release date: | 2010-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of a Polyisoprenoid Binding Domain from Saccharophagus Degradans Implicated in Plant Cell Wall Breakdown
FEBS Lett., 584, 2010
|
|
1WUB
 
 | | Crystal structure of the polyisoprenoid-binding protein, TT1927b, from Thermus thermophilus HB8 | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, conserved hypothetical protein TT1927b | | Authors: | Handa, N, Idaka, M, Terada, T, Hamana, H, Ishizuka, Y, Park, S.-Y, Tame, J.R.H, Doi-Katayama, Y, Hirota, H, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-12-03 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a novel polyisoprenoid-binding protein from Thermus thermophilus HB8
Protein Sci., 14, 2005
|
|
1Y0G
 
 | | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI YCEI PROTEIN, STRUCTURAL GENOMICS | | Descriptor: | 2-[(2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL]PHENOL, Protein yceI | | Authors: | Patskovsky, Y.V, Strokopytov, B, Ramagopal, U, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-11-15 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI YCEI PERIPLASMIC PROTEIN
To be Published
|
|
5W17
 
 | |
5W2V
 
 | |
5W2D
 
 | |
5W30
 
 | | Crystal structure of mutant CJ YCEI protein (CJ-N48C) with monobromobimane guest structure | | Descriptor: | 3-(bromomethyl)-2,5,6-trimethyl-1H,7H-pyrazolo[1,2-a]pyrazole-1,7-dione, Putative periplasmic protein, SULFATE ION, ... | | Authors: | Huber, T.R, Snow, C.D. | | Deposit date: | 2017-06-07 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Installing Guest Molecules at Specific Sites within Scaffold Protein Crystals.
Bioconjug. Chem., 29, 2018
|
|
5W39
 
 | | Crystal structure of mutant CJ YCEI protein (CJ-N182C) with monobromobimane guest structure | | Descriptor: | 3-(bromomethyl)-2,5,6-trimethyl-1H,7H-pyrazolo[1,2-a]pyrazole-1,7-dione, EICOSANE, Polyisoprenoid-binding protein, ... | | Authors: | Huber, T.R, Snow, C.D. | | Deposit date: | 2017-06-07 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Installing Guest Molecules at Specific Sites within Scaffold Protein Crystals.
Bioconjug. Chem., 29, 2018
|
|
5W3A
 
 | |
5W32
 
 | |
5W3C
 
 | |
5W2Z
 
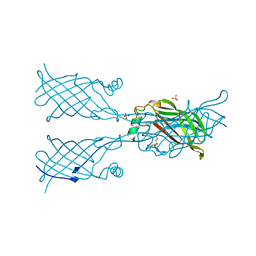 | |
5W2K
 
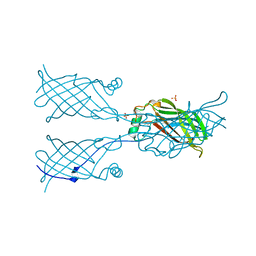 | |
5W2R
 
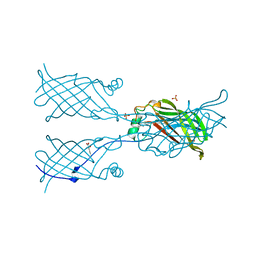 | |
5W2X
 
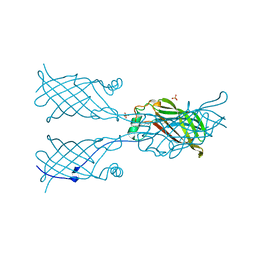 | |
5W31
 
 | |
5W3B
 
 | |
5W37
 
 | |
