8VPO
 
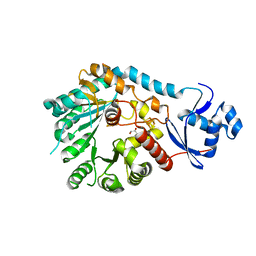 | | X-Ray Crystal Structure of TigE from Paramaledivibacter caminithermalis | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, Radical SAM core domain-containing protein | | Authors: | Grove, T.L, Lachowicz, J.C, Zizola, C. | | Deposit date: | 2024-01-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural, Biochemical, and Bioinformatic Basis for Identifying Radical SAM Cyclopropyl Synthases.
Acs Chem.Biol., 19, 2024
|
|
8VCW
 
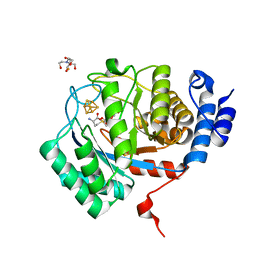 | | X-Ray Crystal Structure of the biotin synthase from B. obeum | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, Biotin synthase, ... | | Authors: | Lachowicz, J.C, Grove, T.L. | | Deposit date: | 2023-12-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery of a Biotin Synthase That Utilizes an Auxiliary 4Fe-5S Cluster for Sulfur Insertion.
J.Am.Chem.Soc., 146, 2024
|
|
8QMN
 
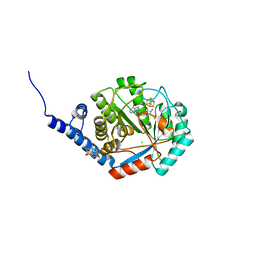 | | [FeFe]-hydrogenase maturase HydE from T. maritima - dialysis experiment - empty structure | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Omeiri, J, Martin, L, Usclat, A, Cherrier, M.V, Nicolet, Y. | | Deposit date: | 2023-09-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Maturation of the [FeFe]-Hydrogenase: Direct Transfer of the ( kappa 3 -cysteinate)Fe II (CN)(CO) 2 Complex B from HydG to HydE.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8QMM
 
 | | M291I variant of the [FeFe]-hydrogenase maturase HydE from Thermotoga maritima | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Omeiri, J, Martin, L, Usclat, A, Cherrier, M.V, Nicolet, Y. | | Deposit date: | 2023-09-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Maturation of the [FeFe]-Hydrogenase: Direct Transfer of the ( kappa 3 -cysteinate)Fe II (CN)(CO) 2 Complex B from HydG to HydE.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8QML
 
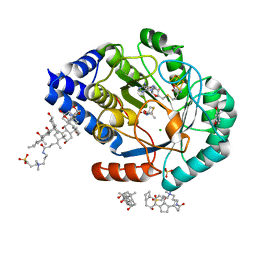 | | (2R,4R)-MeTDA bound HydE structure (control experiment) | | Descriptor: | (2R,4R)-2-methyl-1,3-thiazolidine-2,4-dicarboxylic acid, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, ... | | Authors: | Omeiri, J, Martin, L, Usclat, A, Cherrier, M.V, Nicolet, Y. | | Deposit date: | 2023-09-22 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Maturation of the [FeFe]-Hydrogenase: Direct Transfer of the ( kappa 3 -cysteinate)Fe II (CN)(CO) 2 Complex B from HydG to HydE.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8QMK
 
 | | Enzymatically-produced complex-B bound TmHydE structure | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CARBON MONOXIDE, CHLORIDE ION, ... | | Authors: | Omeiri, J, Martin, L, Usclat, A, Cherrier, M.V, Nicolet, Y. | | Deposit date: | 2023-09-22 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Maturation of the [FeFe]-Hydrogenase: Direct Transfer of the ( kappa 3 -cysteinate)Fe II (CN)(CO) 2 Complex B from HydG to HydE.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8FSI
 
 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to SAM | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, POTASSIUM ION, ... | | Authors: | Moody, J.D, Galambas, A, Lawrence, C.M, Broderick, J.B. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
8FOL
 
 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to SAM, alternate crystal form | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, POTASSIUM ION, ... | | Authors: | Moody, J.D, Saxton, A.J, Galambas, A, Lawrence, C.M, Broderick, J.B. | | Deposit date: | 2022-12-31 | | Release date: | 2023-05-24 | | Last modified: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
8FO0
 
 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to a partially cleaved SAM molecule | | Descriptor: | IRON/SULFUR CLUSTER, POTASSIUM ION, Pyruvate formate-lyase 1-activating enzyme, ... | | Authors: | Moody, J.D, Saxton, A.J, Galambas, A, Lawrence, C.M, Broderick, J.B. | | Deposit date: | 2022-12-29 | | Release date: | 2023-05-24 | | Last modified: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
8AVG
 
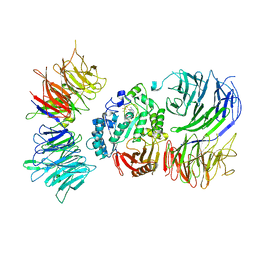 | | Cryo-EM structure of mouse Elp123 with bound SAM | | Descriptor: | Elongator complex protein 1, Elongator complex protein 2, Elongator complex protein 3, ... | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
8ASW
 
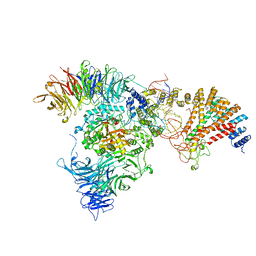 | | Cryo-EM structure of yeast Elp123 in complex with alanine tRNA | | Descriptor: | 5'-DEOXYADENOSINE, Alanine tRNA, Elongator complex protein 1, ... | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-21 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
8ASV
 
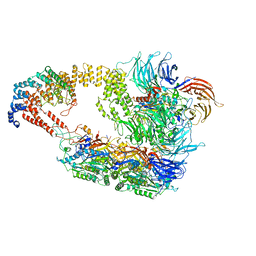 | | Cryo-EM structure of yeast Elongator complex | | Descriptor: | Elongator complex protein 1, Elongator complex protein 2, Elongator complex protein 3, ... | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-21 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
8AI6
 
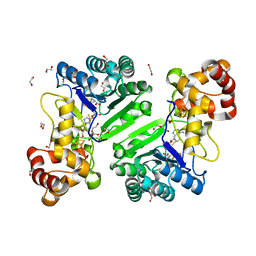 | | Crystal structure of radical SAM epimerase EpeE D210A mutant from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and persulfurated cysteine bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Legrand, P, Fyfe, C.D, Benjdia, A, Berteau, O. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI5
 
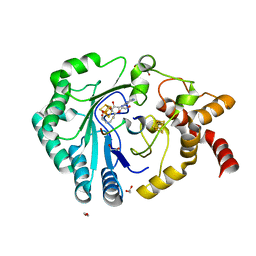 | | Crystal structure of radical SAM epimerase EpeE C223A mutant from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 6 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI4
 
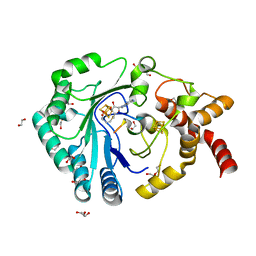 | | Crystal structure of radical SAM epimerase EpeE C223A mutant from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 5 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI3
 
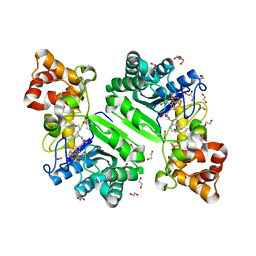 | | Crystal structure of radical SAM epimerase EpeE C223A mutant from Bacillus subtilis with [4Fe-4S] clusters and S-adenosyl-L-methionine bound | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Kubiak, X, Chavas, L.M.G, Legrand, P, Polsinelli, I, Fyfe, C.D, Benjdia, A, Berteau, O. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI2
 
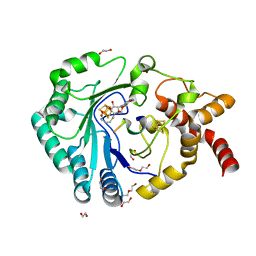 | | Crystal structure of radical SAM epimerase EpeE from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 5 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI1
 
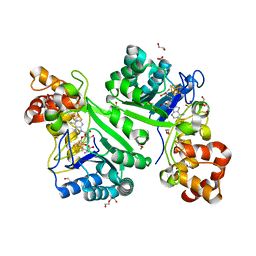 | | Crystal structure of radical SAM epimerase EpeE from Bacillus subtilis with [4Fe-4S] clusters and S-adenosyl-L-homocysteine bound. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Kubiak, X, Polsinelli, I, Chavas, L.M.G, Legrand, P, Fyfe, C.D, Benjdia, A, Berteau, O. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
7X0B
 
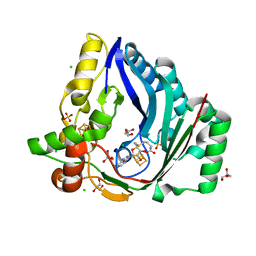 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | 4Fe-4S cluster-binding domain-containing protein, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2022-02-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02027535 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7WZX
 
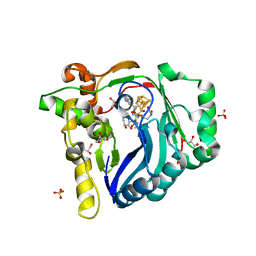 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | (2~{S},4~{S},6~{R})-2-[(2~{S},3~{R},5~{S},6~{R})-3,5-bis(methylamino)-2,4,6-tris(oxidanyl)cyclohexyl]oxy-6-methyl-4-oxidanyl-oxan-3-one, 4Fe-4S cluster-binding domain-containing protein, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2022-02-19 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.980013 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7WZV
 
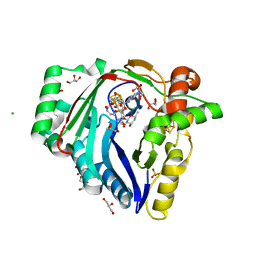 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | (1~{S},2~{R},4~{S},5~{R})-2,4-bis(methylamino)-6-[(2~{S},3~{R},4~{S},6~{R})-6-methyl-3,4-bis(oxidanyl)oxan-2-yl]oxy-cyclohexane-1,3,5-triol, 1,2-ETHANEDIOL, 4Fe-4S cluster-binding domain-containing protein, ... | | Authors: | Zhou, J.H, Hou, X.L. | | Deposit date: | 2022-02-19 | | Release date: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (1.899313 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7VOC
 
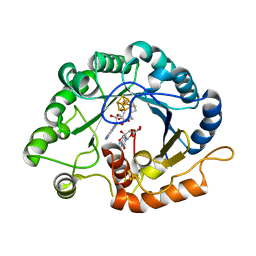 | | The crystal structure of a Radical SAM Enzyme BlsE involved in the Biosynthesis of Blasticidin S | | Descriptor: | (2~{S},3~{S},4~{S},5~{R},6~{R})-6-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4,5-tris(oxidanyl)oxane-2-carboxylic acid, Cytosylglucuronate decarboxylase, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2021-10-13 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.62005424 Å) | | Cite: | Radical S -Adenosyl Methionine Enzyme BlsE Catalyzes a Radical-Mediated 1,2-Diol Dehydration during the Biosynthesis of Blasticidin S.
J.Am.Chem.Soc., 144, 2022
|
|
7VOB
 
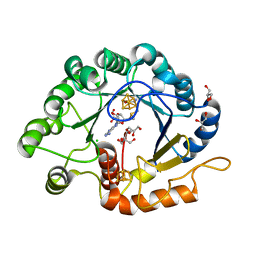 | |
7TOM
 
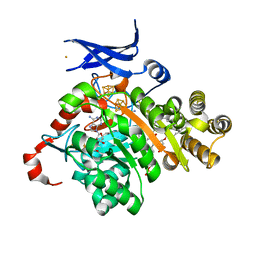 | | X-ray crystal structure of glycerol dibiphytanyl glycerol tetraether - macrocyclic archaeol synthase (GDGT-MAS) from Methanocaldococcus jannaschii with bacterial lipid substrate analog, 5'deoxyadenosine, and methionine bound | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, 5'-DEOXYADENOSINE, FE (III) ION, ... | | Authors: | Lloyd, C.T, Booker, S.J, Boal, A.K. | | Deposit date: | 2022-01-24 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery, structure and mechanism of a tetraether lipid synthase.
Nature, 609, 2022
|
|
7TOL
 
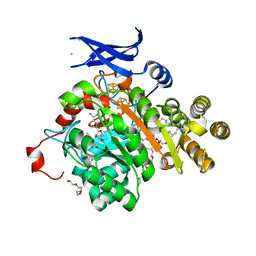 | | X-ray crystal structure of glycerol dibiphytanyl glycerol tetraether - macrocyclic archaeol synthase (GDGT-MAS) from Methanocaldococcus jannaschii with archaeal lipid, 5'deoxyadenosine, and methionine bound | | Descriptor: | 3-PHOSPHORYL-[1,2-DI-PHYTANYL]GLYCEROL, 3-[GLYCEROLYLPHOSPHONYL]-[1,2-DI-PHYTANYL]GLYCEROL, 5'-DEOXYADENOSINE, ... | | Authors: | Lloyd, C.T, Booker, S.J, Boal, A.K. | | Deposit date: | 2022-01-24 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery, structure and mechanism of a tetraether lipid synthase.
Nature, 609, 2022
|
|
