6FVB
 
 | |
6AHO
 
 | | Crystal structure of Kap114p | | Descriptor: | Importin subunit beta-5 | | Authors: | Liao, C.C, Shankar, S, Ahmed, G.R, Hsia, K.C. | | Deposit date: | 2018-08-20 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Karyopherin Kap114p-mediated trans-repression controls ribosomal gene expression under saline stress.
Embo Rep., 21, 2020
|
|
3GB8
 
 | | Crystal structure of CRM1/Snurportin-1 complex | | Descriptor: | Exportin-1, Snurportin-1 | | Authors: | Dong, X, Biswas, A, Suel, K.E, Jackson, L.K, Martinez, R, Gu, H, Chook, Y.M. | | Deposit date: | 2009-02-19 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for leucine-rich nuclear export signal recognition by CRM1.
Nature, 458, 2009
|
|
4XRI
 
 | | Crystal Structure of Importin Beta in an Ammonium Sulfate Condition | | Descriptor: | GLYCEROL, Putative uncharacterized protein, SULFATE ION | | Authors: | Tauchert, M.J, Neumann, P, Ficner, R, Dickmanns, A. | | Deposit date: | 2015-01-21 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Impact of the crystallization condition on importin-beta conformation.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4XRK
 
 | |
1GCJ
 
 | | N-TERMINAL FRAGMENT OF IMPORTIN-BETA | | Descriptor: | IMPORTIN BETA | | Authors: | Lee, S.J, Imamoto, N, Sakai, H, Nakagawa, A, Kose, S, Koike, M, Yamamoto, M, Kumasaka, T, Yoneda, Y, Tsukihara, T. | | Deposit date: | 2000-07-31 | | Release date: | 2000-10-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The adoption of a twisted structure of importin-beta is essential for the protein-protein interaction required for nuclear transport.
J.Mol.Biol., 302, 2000
|
|
3ZKV
 
 | |
4BSN
 
 | | Crystal structure of the Nuclear Export Receptor CRM1 (exportin-1) lacking the C-terminal helical extension at 4.1A | | Descriptor: | EXPORTIN-1 | | Authors: | Dian, C, Bernaudat, F, Langer, K, Oliva, M.F, Fornerod, M, Schoehn, G, Muller, C.W, Petosa, C. | | Deposit date: | 2013-06-11 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structure of a Truncation Mutant of the Nuclear Export Factor Crm1 Provides Insights Into the Auto-Inhibitory Role of its C-Terminal Helix.
Structure, 21, 2013
|
|
4BSM
 
 | | Crystal structure of the Nuclear Export Receptor CRM1 (exportin-1) lacking the C-terminal helical extension at 4.5A | | Descriptor: | EXPORTIN-1 | | Authors: | Dian, C, Bernaudat, F, Langer, K, Oliva, M.F, Fornerod, M, Schoehn, G, Muller, C.W, Petosa, C. | | Deposit date: | 2013-06-10 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structure of a Truncation Mutant of the Nuclear Export Factor Crm1 Provides Insights Into the Auto-Inhibitory Role of its C-Terminal Helix.
Structure, 21, 2013
|
|
3VYC
 
 | |
5YU7
 
 | |
5OWU
 
 | | Kap95:Nup1 complex | | Descriptor: | Importin subunit beta-1, Nucleoporin NUP1 | | Authors: | Stewart, M. | | Deposit date: | 2017-09-04 | | Release date: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the high-affinity binding of nucleoporin Nup1p to the Saccharomyces cerevisiae importin-beta homologue, Kap95p.
J. Mol. Biol., 349, 2005
|
|
2H4M
 
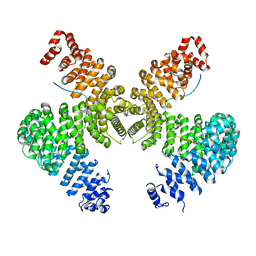 | | Karyopherin Beta2/Transportin-M9NLS | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, Transportin-1 | | Authors: | Chook, Y.M, Cansizoglu, A.E. | | Deposit date: | 2006-05-24 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Rules for nuclear localization sequence recognition by karyopherin beta 2.
Cell(Cambridge,Mass.), 126, 2006
|
|
7YPZ
 
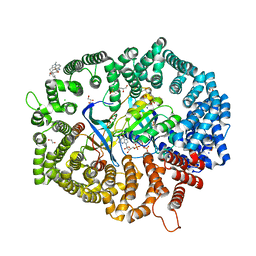 | | Zafirlukast in complex with CRM1-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Searching for Novel Noncovalent Nuclear Export Inhibitors through a Drug Repurposing Approach.
J.Med.Chem., 66, 2023
|
|
8DYO
 
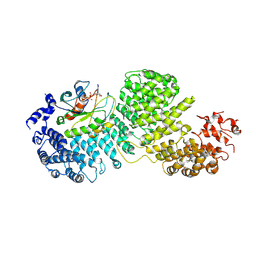 | | Cryo-EM structure of Importin-4 bound to RanGTP | | Descriptor: | GTP-binding nuclear protein GSP1/CNR1, GUANOSINE-5'-TRIPHOSPHATE, Importin-4, ... | | Authors: | Bernardes, N.E, Fung, H.Y.J, Li, Y, Chen, Z, Chook, Y.M. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structure of IMPORTIN-4 bound to the H3-H4-ASF1 histone-histone chaperone complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7L5E
 
 | |
6TVO
 
 | | Human CRM1-RanGTP in complex with Leptomycin B | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shaikhqasem, A, Ficner, R. | | Deposit date: | 2020-01-10 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Characterization of Inhibition Reveals Distinctive Properties for Human andSaccharomyces cerevisiaeCRM1.
J.Med.Chem., 63, 2020
|
|
3EA5
 
 | | Kap95p Binding Induces the Switch Loops of RanGDP to adopt the GTP-bound Conformation: Implications for Nuclear Import Complex Assembly Dynamics | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, Importin subunit beta-1, ... | | Authors: | Forwood, J.K, Lonhienne, J.K, Guncar, G, Stewart, M, Marfori, M, Kobe, B. | | Deposit date: | 2008-08-24 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kap95p binding induces the switch loops of RanGDP to adopt the GTP-bound conformation: implications for nuclear import complex assembly dynamics.
J.Mol.Biol., 383, 2008
|
|
4OO6
 
 | | Crystal structure of human KAP-beta2 bound to the NLS of HCC1 (Hepato Cellular Carcinoma protein 1) | | Descriptor: | RNA-binding protein 39, Transportin-1 | | Authors: | Sampathkumar, P, Brower, A, Soniat, M, Bonanno, J, Hillerich, B, Seidel, R.D, Rout, M.P, Chook, Y.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2014-01-30 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human KAP-beta2 bound to the NLS of HCC1 (Hepato Cellular Carcinoma protein 1)
to be published
|
|
6N89
 
 | |
6N1Z
 
 | | Importin-9 bound to H2A-H2B | | Descriptor: | Histone H2A, Histone H2B 1.1, Importin-9 | | Authors: | Tomchick, D.R, Chook, Y.M, Padavannil, A. | | Deposit date: | 2018-11-12 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Importin-9 wraps around the H2A-H2B core to act as nuclear importer and histone chaperone.
Elife, 8, 2019
|
|
1Z3H
 
 | | The exportin Cse1 in its cargo-free, cytoplasmic state | | Descriptor: | Importin alpha re-exporter, MAGNESIUM ION | | Authors: | Cook, A, Fernandez, E, Lindner, D, Ebert, J, Schlenstedt, G, Conti, E. | | Deposit date: | 2005-03-12 | | Release date: | 2005-05-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of the nuclear export receptor cse1 in its cytosolic state reveals a closed conformation incompatible with cargo binding
Mol.Cell, 18, 2005
|
|
3GJX
 
 | | Crystal Structure of the Nuclear Export Complex CRM1-Snurportin1-RanGTP | | Descriptor: | CHLORIDE ION, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Monecke, T, Guettler, T, Neumann, P, Dickmanns, A, Goerlich, D, Ficner, R. | | Deposit date: | 2009-03-09 | | Release date: | 2009-05-26 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Nuclear Export Receptor CRM1 in Complex with Snurportin1 and RanGTP.
Science, 2009
|
|
1IBR
 
 | | COMPLEX OF RAN WITH IMPORTIN BETA | | Descriptor: | GTP-binding nuclear protein RAN, Importin beta-1 subunit, MAGNESIUM ION, ... | | Authors: | Vetter, I.R, Arndt, A, Kutay, U, Goerlich, D, Wittinghofer, A. | | Deposit date: | 1999-05-14 | | Release date: | 1999-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural view of the Ran-Importin beta interaction at 2.3 A resolution
Cell(Cambridge,Mass.), 97, 1999
|
|
1UKL
 
 | | Crystal structure of Importin-beta and SREBP-2 complex | | Descriptor: | Importin beta-1 subunit, Sterol regulatory element binding protein-2 | | Authors: | Lee, S.J, Sekimoto, T, Yamashita, E, Nagoshi, E, Nakagawa, A, Imamoto, N, Yoshimura, M, Sakai, H, Tsukihara, T, Yoneda, Y. | | Deposit date: | 2003-08-26 | | Release date: | 2003-12-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Importin-beta Bound to SREBP-2: Nuclear Import of a Transcription Factor
Science, 302, 2003
|
|
