6TYU
 
 | |
6TYZ
 
 | |
6TYW
 
 | |
6TYX
 
 | |
6TYV
 
 | |
5Y59
 
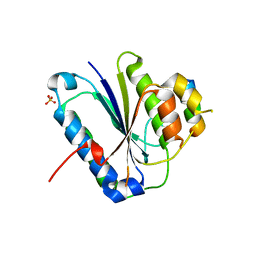 | | Crystal structure of Ku80 and Sir4 | | Descriptor: | ATP-dependent DNA helicase II subunit 2, SULFATE ION, Sir4p | | Authors: | Chen, H, Xue, J, Wu, J, Lei, M. | | Deposit date: | 2017-08-08 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structural Insights into Yeast Telomerase Recruitment to Telomeres
Cell, 172, 2018
|
|
6TYT
 
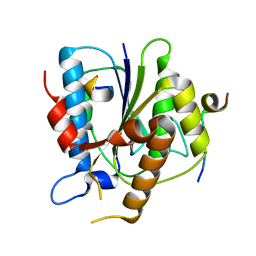 | |
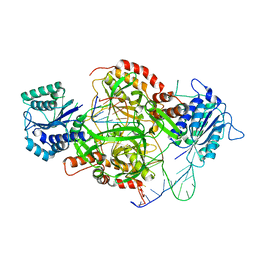
8AG4
 
 | | Vaccinia C16 protein bound to Ku70/Ku80 | | Descriptor: | Protein C10, X-ray repair cross-complementing protein 5, X-ray repair cross-complementing protein 6 | | Authors: | Rivera-Calzada, A, Arribas-Bosacoma, R, Pearl, L.H, Llorca, O. | | Deposit date: | 2022-07-19 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural basis for the inactivation of cytosolic DNA sensing by the vaccinia virus.
Nat Commun, 13, 2022
|
|
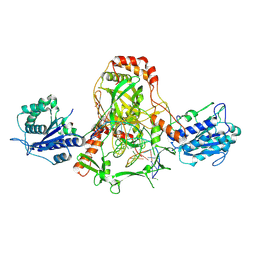
1JEY
 
 | | Crystal Structure of the Ku heterodimer bound to DNA | | Descriptor: | DNA (34-MER), DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*TP*TP*GP*GP*GP*CP*GP*CP*G)-3'), Ku70, ... | | Authors: | Walker, J.R, Corpina, R.A, Goldberg, J. | | Deposit date: | 2001-06-19 | | Release date: | 2001-08-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair.
Nature, 412, 2001
|
|
7ZYG
 
 | | CryoEM structure of Ku heterodimer bound to DNA, PAXX and XLF | | Descriptor: | DNA, Non-homologous end-joining factor 1, PHOSPHATE ION, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
1JEQ
 
 | |
7ZVT
 
 | | CryoEM structure of Ku heterodimer bound to DNA | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*TP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-17 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
5Y58
 
 | | Crystal structure of Ku70/80 and TLC1 | | Descriptor: | ATP-dependent DNA helicase II subunit 1, ATP-dependent DNA helicase II subunit 2, TLC1 | | Authors: | Chen, H, Xue, J, Wu, J, Lei, M. | | Deposit date: | 2017-08-08 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into Yeast Telomerase Recruitment to Telomeres
Cell, 172, 2018
|
|
7ZWA
 
 | | CryoEM structure of Ku heterodimer bound to DNA and PAXX | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*T)-3'), PHOSPHATE ION, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
6ERH
 
 | | Complex of XLF and heterodimer Ku bound to DNA | | Descriptor: | DNA (21-MER), DNA (34-MER), Non-homologous end-joining factor 1, ... | | Authors: | Nemoz, C, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2017-10-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6ERG
 
 | | Complex of XLF and heterodimer Ku bound to DNA | | Descriptor: | DNA (21-MER), DNA (34-MER), Non-homologous end-joining factor 1, ... | | Authors: | Nemoz, C, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2017-10-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7Z87
 
 | | DNA-PK in the active state | | Descriptor: | (~{S})-[2-chloranyl-4-fluoranyl-5-(7-morpholin-4-ylquinazolin-4-yl)phenyl]-(6-methoxypyridazin-3-yl)methanol, DNA (26-MER), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Liang, S, Blundell, T.L. | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-18 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Human DNA-dependent protein kinase activation mechanism.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8ASC
 
 | | Ku70/80 binds to the Ku-binding motif of PAXX | | Descriptor: | DNA (5'-D(P*CP*GP*GP*AP*TP*CP*GP*AP*GP*GP*GP*CP*CP*CP*GP*AP*TP*AP*T)-3'), DNA (5'-D(P*GP*GP*GP*CP*CP*CP*TP*CP*GP*AP*TP*CP*CP*G)-3'), Protein PAXX, ... | | Authors: | Seif El Dahan, M, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
7SGL
 
 | | DNA-PK complex of DNA end processing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-dependent protein kinase catalytic subunit, Hairpin_1, ... | | Authors: | Liu, L, Li, J, Chen, X, Yang, W, Gellert, M. | | Deposit date: | 2021-10-06 | | Release date: | 2022-01-12 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
6ERF
 
 | | Complex of APLF factor and Ku heterodimer bound to DNA | | Descriptor: | Aprataxin and PNK-like factor, DNA (34-MER), DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*TP*TP*GP*GP*GP*CP*GP*CP*G)-3'), ... | | Authors: | Nemoz, C, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2017-10-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining.
Nat.Struct.Mol.Biol., 25, 2018
|
|
7AXZ
 
 | | Ku70/80 complex apo form | | Descriptor: | X-ray repair cross-complementing protein 5, X-ray repair cross-complementing protein 6 | | Authors: | Hnizda, A, Tesina, P, Novak, P, Blundell, T.L. | | Deposit date: | 2020-11-10 | | Release date: | 2021-02-10 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | SAP domain forms a flexible part of DNA aperture in Ku70/80.
Febs J., 288, 2021
|
|
7SU3
 
 | | CryoEM structure of DNA-PK complex VII | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), ... | | Authors: | Chen, X, Liu, L, Gellert, M, Yang, W. | | Deposit date: | 2021-11-16 | | Release date: | 2022-01-12 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
7Z88
 
 | | DNA-PK in the intermediate state | | Descriptor: | (~{S})-[2-chloranyl-4-fluoranyl-5-(7-morpholin-4-ylquinazolin-4-yl)phenyl]-(6-methoxypyridazin-3-yl)methanol, DNA (26-MER), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Liang, S, Blundell, T.L. | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-18 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Human DNA-dependent protein kinase activation mechanism.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AG5
 
 | | Vaccinia C16 protein bound to Ku70/Ku80 | | Descriptor: | Ku70-Xrcc6, Protein C10, X-ray repair cross-complementing protein 5 | | Authors: | Rivera-Calzada, A, Arribas-Bosacoma, R, Pearl, L.H, Llorca, O. | | Deposit date: | 2022-07-19 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis for the inactivation of cytosolic DNA sensing by the vaccinia virus.
Nat Commun, 13, 2022
|
|
7ZT6
 
 | |
