5TT1
 
 | |
4YE5
 
 | | The crystal structure of a peptidoglycan synthetase from Bifidobacterium adolescentis ATCC 15703 | | Descriptor: | ACETATE ION, GLYCEROL, Peptidoglycan synthetase penicillin-binding protein 3 | | Authors: | Cuff, M, Tan, K, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-18 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | The crystal structure of a peptidoglycan synthetase from Bifidobacterium adolescentis ATCC 15703
To Be Published
|
|
1VQQ
 
 | |
1K25
 
 | |
7ZG8
 
 | |
8TJ3
 
 | |
8U55
 
 | |
4FSF
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 complexed with compound 14 | | Descriptor: | (4R,5S,8Z)-8-(2-amino-1,3-thiazol-4-yl)-1-[3-(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)-1,2-oxazol-5-yl]-5-formyl-11,11-dimethyl-1,7-dioxo-4-(sulfoamino)-10-oxa-2,6,9-triazadodec-8-en-12-oic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel monobactams utilizing a siderophore uptake mechanism for the treatment of gram-negative infections.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3EQU
 
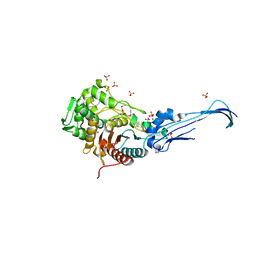 | | Crystal structure of penicillin-binding protein 2 from Neisseria gonorrhoeae | | Descriptor: | GLYCEROL, Penicillin-binding protein 2, SULFATE ION | | Authors: | Powell, A.J, Deacon, A.M, Nicholas, R.A, Davies, C. | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Penicillin-binding Protein 2 from Penicillin-susceptible and -resistant Strains of Neisseria gonorrhoeae Reveal an Unexpectedly Subtle Mechanism for Antibiotic Resistance.
J.Biol.Chem., 284, 2009
|
|
3EQV
 
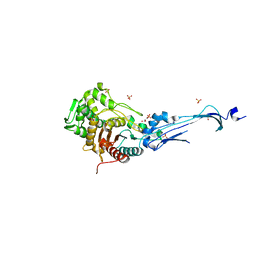 | | Crystal structure of penicillin-binding protein 2 from Neisseria gonorrhoeae containing four mutations associated with penicillin resistance | | Descriptor: | GLYCEROL, Penicillin-binding protein 2, SULFATE ION | | Authors: | Powell, A.J, Deacon, A.M, Nicholas, R.A, Davies, C. | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Penicillin-binding Protein 2 from Penicillin-susceptible and -resistant Strains of Neisseria gonorrhoeae Reveal an Unexpectedly Subtle Mechanism for Antibiotic Resistance.
J.Biol.Chem., 284, 2009
|
|
4KQQ
 
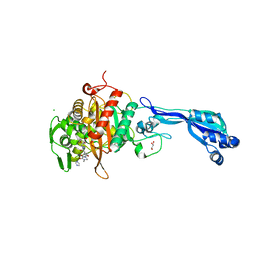 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH (5S)-Penicilloic Acid | | Descriptor: | (2S,4S)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
6XV5
 
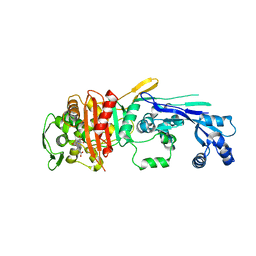 | |
6Y6Z
 
 | | Structure of Pseudomonas aeruginosa Penicillin-Binding Protein 3 (PBP3) in complex with Compound 1 | | Descriptor: | GLYCEROL, Peptidoglycan D,D-transpeptidase FtsI, ~{tert}-butyl ~{N}-[(2~{S})-2-methyl-4-oxidanyl-1-oxidanylidene-pent-4-en-2-yl]carbamate | | Authors: | Newman, H, Bellini, D, Dowson, C.G. | | Deposit date: | 2020-02-27 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion.
Chem Sci, 11, 2020
|
|
4WEJ
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 with a R4 substituted allyl monocarbam | | Descriptor: | (3R,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-4-formyl-1-[({3-[(5R)-5-hydroxy-4-oxo-4,5-dihydropyridin-2-yl]-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-10,10-dimethyl-1,6-dioxo-3-(prop-2-en-1-yl)-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.045 Å) | | Cite: | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
4WEL
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 with SMC-3176 | | Descriptor: | (3S,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-4-formyl-1-[({3-(5-hydroxy-4-oxo-3,4-dihydropyridin-2-yl)-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-3,10,10-trimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
4WEK
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 with a R4 substituted vinyl monocarbam | | Descriptor: | (3S,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-3-ethenyl-4-formyl-1-[({3-(5-hydroxy-4-oxo-3,4-dihydropyridin-2-yl)-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-10,10-dimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
6Y6U
 
 | | Structure of Pseudomonas aeruginosa Penicillin-Binding Protein 3 (PBP3) in complex with Compound 6 | | Descriptor: | 2-(4-hydroxyphenyl)-~{N}-[(2~{S})-2-methyl-4-oxidanyl-1-oxidanylidene-pent-4-en-2-yl]ethanamide, GLYCEROL, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Newman, H, Bellini, D, Dowson, C.G. | | Deposit date: | 2020-02-27 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion
Chem Sci, 11, 2020
|
|
6PL5
 
 | | Structural coordination of polymerization and crosslinking by a peptidoglycan synthase complex | | Descriptor: | Penicillin-binding protein 2/cell division protein FtsI, Peptidoglycan glycosyltransferase RodA, Unknown peptide | | Authors: | Sjodt, M, Rohs, P.D.A, Erlandson, S.C, Zheng, S, Rudner, D.Z, Bernhardt, T.G, Kruse, A.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex.
Nat Microbiol, 5, 2020
|
|
6PL6
 
 | | Structural coordination of polymerization and crosslinking by a peptidoglycan synthase complex | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 2/cell division protein FtsI, Peptidoglycan glycosyltransferase RodA, ... | | Authors: | Sjodt, M, Rohs, P.D.A, Erlandson, S.C, Zheng, S, Rudner, D.Z, Bernhardt, T.G, Kruse, A.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex.
Nat Microbiol, 5, 2020
|
|
6Q9N
 
 | | Crystal structure of PBP2a from MRSA in complex with piperacillin and quinazolinone | | Descriptor: | 3-[2-[(~{E})-2-(4-ethynylphenyl)ethenyl]-4-oxidanylidene-quinazolin-3-yl]benzoic acid, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Martinez-Caballero, S, Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2018-12-18 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Quinazolinone Allosteric Inhibitor of PBP 2a Synergizes with Piperacillin and Tazobactam against Methicillin-Resistant Staphylococcus aureus.
Antimicrob.Agents Chemother., 63, 2019
|
|
6BSR
 
 | | Crystal structure of penicillin-binding protein 4 (PBP4) from Enterococcus faecalis in the benzylpenicillin bound form. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Moon, T.M, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6C84
 
 | |
6BSQ
 
 | | Enterococcus faecalis Penicillin Binding Protein 4 (PBP4) | | Descriptor: | CHLORIDE ION, GLYCEROL, PBP4 protein | | Authors: | Moon, T.M, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
4KQO
 
 | | Crystal structure of penicillin-binding protein 3 from pseudomonas aeruginosa in complex with piperacillin | | Descriptor: | CHLORIDE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
4KQR
 
 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH (5S)-Penicilloic Acid | | Descriptor: | (2S,4S)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
