5FTW
 
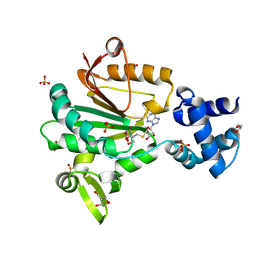 | | Crystal structure of glutamate O-methyltransferase in complex with S- adenosyl-L-homocysteine (SAH) from Bacillus subtilis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHEMOTAXIS PROTEIN METHYLTRANSFERASE, GLYCEROL, ... | | Authors: | Sharma, R, Dhindwal, S, Batra, M, Aggarwal, M, Kumar, P, Tomar, S. | | Deposit date: | 2016-01-18 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Pentapeptide-Independent Chemotaxis Receptor Methyltransferase (Cher) Reveals Idiosyncratic Structural Determinants for Receptor Recognition.
J.Struct.Biol., 196, 2016
|
|
1AF7
 
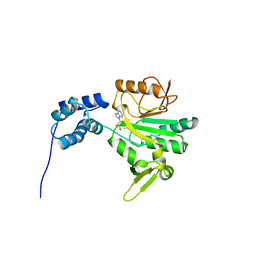 | | CHER FROM SALMONELLA TYPHIMURIUM | | Descriptor: | CHEMOTAXIS RECEPTOR METHYLTRANSFERASE CHER, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Djordjevic, S, Stock, A.M. | | Deposit date: | 1997-03-22 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motif for binding S-adenosylmethionine.
Structure, 5, 1997
|
|
1BC5
 
 | |
5XLX
 
 | |
5XLY
 
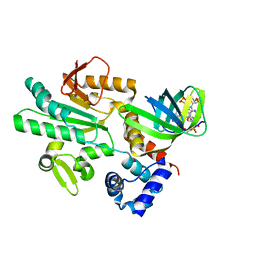 | | Crystal structure of CheR1 in complex with c-di-GMP-bound MapZ | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Chemotaxis protein methyltransferase 1, Cyclic diguanosine monophosphate-binding protein PA4608 | | Authors: | Yuan, Z, Zhu, Y, Gu, L. | | Deposit date: | 2017-05-12 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Structural basis for the regulation of chemotaxis by MapZ in the presence of c-di-GMP
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5Y4S
 
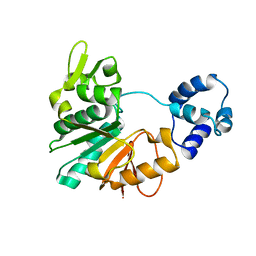 | | Structure of a methyltransferase complex | | Descriptor: | Chemotaxis protein methyltransferase 1 | | Authors: | Yan, X, Xin, L, Tan, Y.J, Jin, S, Liang, Z.X, Gao, Y.G. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.405 Å) | | Cite: | Structural analyses unravel the molecular mechanism of cyclic di-GMP regulation of bacterial chemotaxis via a PilZ adaptor protein.
J. Biol. Chem., 293, 2018
|
|
5Y4R
 
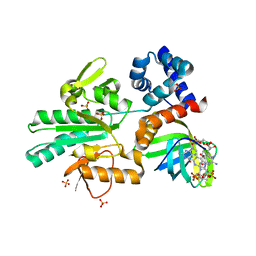 | | Structure of a methyltransferase complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Chemotaxis protein methyltransferase 1, Cyclic diguanosine monophosphate-binding protein PA4608, ... | | Authors: | Yan, X, Xin, L, Tan, Y.J, Jin, S, Liang, Z.X, Gao, Y.G. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Structural analyses unravel the molecular mechanism of cyclic di-GMP regulation of bacterial chemotaxis via a PilZ adaptor protein.
J. Biol. Chem., 293, 2018
|
|
