8SPX
 
 | | PS3 F1 Rotorless, high ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2023-05-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | The series of conformational states adopted by rotorless F 1 -ATPase during its hydrolysis cycle.
Structure, 32, 2024
|
|
8SPW
 
 | | PS3 F1 Rotorless, low ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2023-05-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The series of conformational states adopted by rotorless F 1 -ATPase during its hydrolysis cycle.
Structure, 32, 2024
|
|
8SPV
 
 | | PS3 F1 Rotorless, no ATP | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2023-05-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | The series of conformational states adopted by rotorless F 1 -ATPase during its hydrolysis cycle.
Structure, 32, 2024
|
|
8IGW
 
 | | Hexameric Ring Complex of Engineered V1-ATPase bound to 4 ADPs: A3(De)3_(ADP)3cat,1non-cat, Hexameric Ring Complex of Engineered V1-ATPase bound to 5 ADPs: A3(De)3_(ADP)3cat,2non-cat | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, V-type sodium ATPase catalytic subunit A, ... | | Authors: | Kosugi, T, Tanabe, M, Koga, N. | | Deposit date: | 2023-02-21 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Design of allosteric sites into rotary motor V 1 -ATPase by restoring lost function of pseudo-active sites.
Nat.Chem., 15, 2023
|
|
8IGV
 
 | | Hexameric Ring Complex of Engineered V1-ATPase bound to 5 ADPs: A3(De)3_(ADP-Pi)1cat(ADP)2cat,2non-cat | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kosugi, T, Tanabe, M, Koga, N. | | Deposit date: | 2023-02-21 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Design of allosteric sites into rotary motor V 1 -ATPase by restoring lost function of pseudo-active sites.
Nat.Chem., 15, 2023
|
|
8IGU
 
 | |
8G0E
 
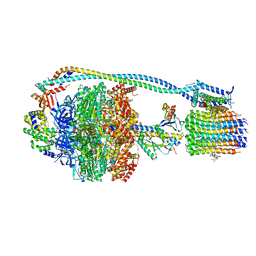 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 3 | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
8G0C
 
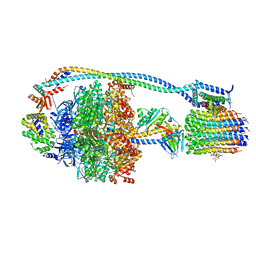 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 1 (backbone model) | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
8G09
 
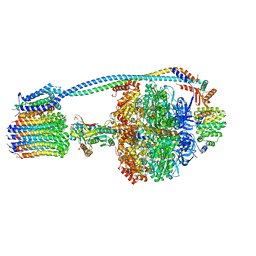 | |
8G0A
 
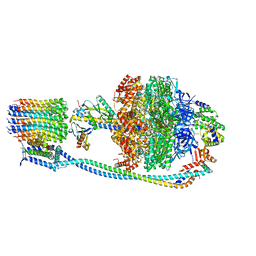 | |
8G08
 
 | |
8G0D
 
 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 2 (backbone model) | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
8FKJ
 
 | | Yeast ATP Synthase in conformation-3, at pH 6 | | Descriptor: | ATP synthase protein 8, ATP synthase subunit 4, mitochondrial, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8FL8
 
 | | Yeast ATP Synthase structure in presence of MgATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase protein 8, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8HH3
 
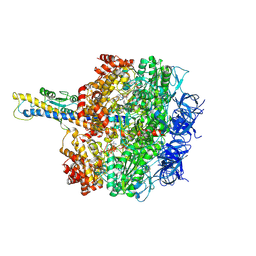 | | F1 domain of FoF1-ATPase from Bacillus PS3,90 degrees,highATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH1
 
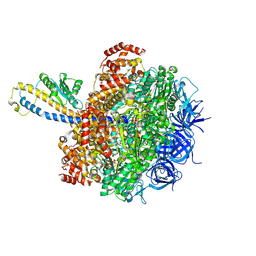 | | FoF1-ATPase from Bacillus PS3, 81 degrees, highATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH7
 
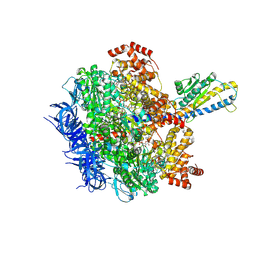 | | F1 domain of FoF1-ATPase from Bacillus PS3, 81 degrees, lowATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH4
 
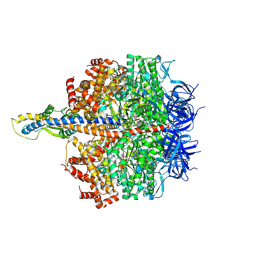 | | F1 domain of FoF1-ATPase from Bacillus PS3,101 degrees, highATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH5
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,120 degrees,highATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HHC
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,post-hyd',lowATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH8
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,post-hyd,lowATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH2
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,post-hyd,highATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HHB
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,step waiting,lowATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HHA
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,120 degrees,lowATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH6
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,step waiting,highATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
