7U58
 
 | |
6PEU
 
 | |
6PE3
 
 | |
6GRG
 
 | | E. coli Microcin synthetase McbBCD complex with pro-MccB17, ADP and phosphate bound | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ghilarov, D, Stevenson, C.E.M, Travin, D.Y, Piskunova, J, Serebryakova, M, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2018-06-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Architecture of Microcin B17 Synthetase: An Octameric Protein Complex Converting a Ribosomally Synthesized Peptide into a DNA Gyrase Poison.
Mol. Cell, 73, 2019
|
|
6GRI
 
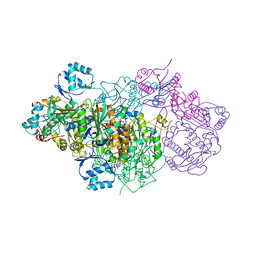 | | E. coli Microcin synthetase McbBCD complex | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, Microcin B17-processing protein McbB, ... | | Authors: | Ghilarov, D, Stevenson, C.E.M, Travin, D.Y, Piskunova, J, Serebryakova, M, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2018-06-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Architecture of Microcin B17 Synthetase: An Octameric Protein Complex Converting a Ribosomally Synthesized Peptide into a DNA Gyrase Poison.
Mol. Cell, 73, 2019
|
|
6GRH
 
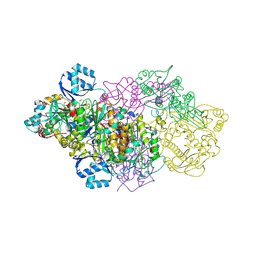 | | E. coli Microcin synthetase McbBCD complex with truncated pro-MccB17 bound | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin microcin B17, CHLORIDE ION, ... | | Authors: | Ghilarov, D, Stevenson, C.E.M, Travin, D.Y, Piskunova, J, Serebryakova, M, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2018-06-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Architecture of Microcin B17 Synthetase: An Octameric Protein Complex Converting a Ribosomally Synthesized Peptide into a DNA Gyrase Poison.
Mol. Cell, 73, 2019
|
|
6GOS
 
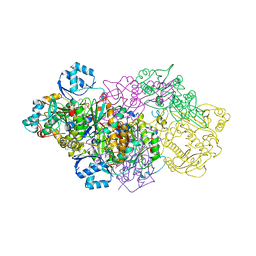 | | E. coli Microcin synthetase McbBCD complex with pro-MccB17 bound | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin microcin B17, CHLORIDE ION, ... | | Authors: | Ghilarov, D, Stevenson, C.E.M, Travin, D.Y, Piskunova, J, Serebryakova, M, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2018-06-04 | | Release date: | 2019-01-30 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Architecture of Microcin B17 Synthetase: An Octameric Protein Complex Converting a Ribosomally Synthesized Peptide into a DNA Gyrase Poison.
Mol. Cell, 73, 2019
|
|
6CIB
 
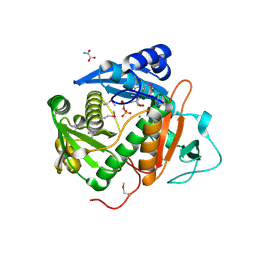 | |
6CI7
 
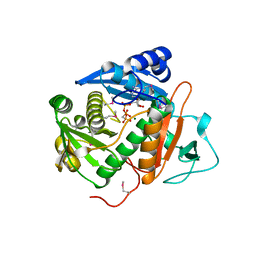 | |
4V1V
 
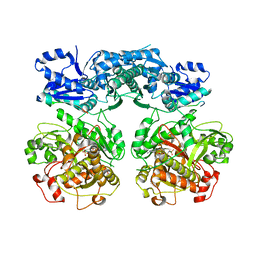 | | Heterocyclase in complex with substrate and Cofactor | | Descriptor: | LYND, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Koehnke, J, Naismith, J.H. | | Deposit date: | 2014-10-02 | | Release date: | 2015-01-14 | | Last modified: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural Analysis of Leader Peptide Binding Enables Leader-Free Cyanobactin Processing.
Nat.Chem.Biol., 11, 2015
|
|
4V1T
 
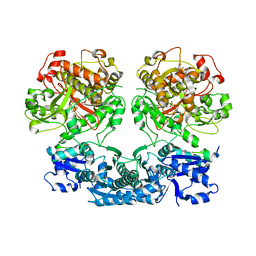 | |
4V1U
 
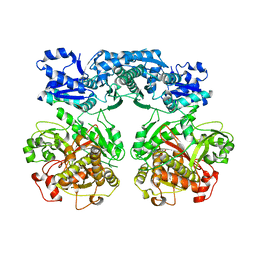 | |
4Q85
 
 | | YcaO with Non-hydrolyzable ATP (AMPCPP) Bound | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, Ribosomal protein S12 methylthiotransferase accessory factor YcaO | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2014-04-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Discovery of a new ATP-binding motif involved in peptidic azoline biosynthesis.
Nat.Chem.Biol., 10, 2014
|
|
4Q86
 
 | | YcaO with AMP Bound | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Ribosomal protein S12 methylthiotransferase accessory factor YcaO | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2014-04-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of a new ATP-binding motif involved in peptidic azoline biosynthesis.
Nat.Chem.Biol., 10, 2014
|
|
4Q84
 
 | | Apo YcaO | | Descriptor: | MERCURY (II) ION, Ribosomal protein S12 methylthiotransferase accessory factor YcaO | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2014-04-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of a new ATP-binding motif involved in peptidic azoline biosynthesis.
Nat.Chem.Biol., 10, 2014
|
|
4BS9
 
 | | Structure of the heterocyclase TruD | | Descriptor: | TRUD, ZINC ION | | Authors: | Koehnke, J, Bent, A.F, Zollman, D, Smith, K, Houssen, W.E, Zhu, X, Mann, G, Lebl, T, Scharff, R, Shirran, S, Botting, C.H, Jaspars, M, Schwarz-Linek, U, Naismith, J.H. | | Deposit date: | 2013-06-09 | | Release date: | 2013-11-06 | | Last modified: | 2014-01-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Cyanobactin Heterocyclase Enzyme: A Processive Adenylase that Operates with a Defined Order of Reaction.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
