5C8A
 
 | |
8C76
 
 | | Light-state 2.5 Angstrom wild-type X-ray crystal structure of the cobalamin binding domain belonging to a light-dependent transcription regulator TtCarH obtained under aerobic conditions from a form 2 crystal illuminated during 5 s | | Descriptor: | COBALAMIN, Probable transcriptional regulator | | Authors: | Rios-Santacruz, R, Colletier, J.P, Schiro, G, Weik, M. | | Deposit date: | 2023-01-12 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Redox driven B 12 -ligand switch drives CarH photoresponse.
Nat Commun, 14, 2023
|
|
8C73
 
 | | Dark state 1.8 Angstrom crystal structure of cobalamin binding domain belonging to a light-dependent transcription regulator TtCarH obtained under aerobic condition form ll | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Probable transcriptional regulator | | Authors: | Rios-Santacruz, R, Colletier, J.P, Schiro, G, Weik, M. | | Deposit date: | 2023-01-12 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Redox driven B 12 -ligand switch drives CarH photoresponse.
Nat Commun, 14, 2023
|
|
8C36
 
 | |
8C33
 
 | |
8C34
 
 | |
8C35
 
 | |
8C37
 
 | |
8C31
 
 | |
8C32
 
 | |
3WHP
 
 | | Crystal structure of the C-terminal domain of Themus thermophilus LitR in complex with cobalamin | | Descriptor: | COBALAMIN, Probable transcriptional regulator | | Authors: | Agari, Y, Takano, H, Beppu, T, Ueda, K, Shinkai, A. | | Deposit date: | 2013-08-30 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of the C-terminal domain of Themus thermophilus LitR in complex with cobalamin
To be Published
|
|
5C8F
 
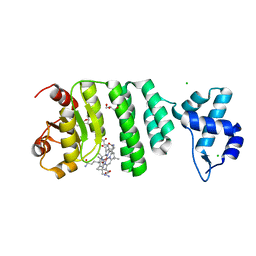 | |
5C8E
 
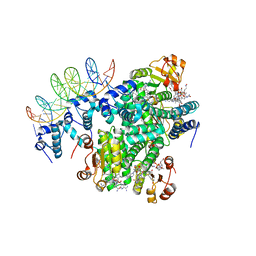 | |
5C8D
 
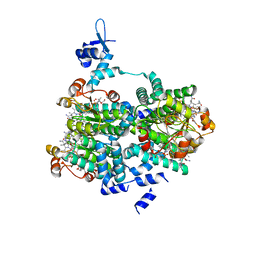 | |
1BMT
 
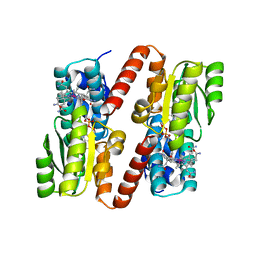 | | HOW A PROTEIN BINDS B12: A 3.O ANGSTROM X-RAY STRUCTURE OF THE B12-BINDING DOMAINS OF METHIONINE SYNTHASE | | Descriptor: | CO-METHYLCOBALAMIN, METHIONINE SYNTHASE | | Authors: | Drennan, C.L, Huang, S, Drummond, J.T, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 1994-09-02 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How a protein binds B12: A 3.0 A X-ray structure of B12-binding domains of methionine synthase.
Science, 266, 1994
|
|
2I2X
 
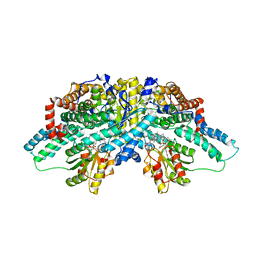 | | Crystal structure of methanol:cobalamin methyltransferase complex MtaBC from Methanosarcina barkeri | | Descriptor: | 5-HYDROXYBENZIMIDAZOLYLCOB(III)AMIDE, Methyltransferase 1, POTASSIUM ION, ... | | Authors: | Hagemeier, C.H, Kruer, M, Thauer, R.K, Warkentin, E, Ermler, U. | | Deposit date: | 2006-08-17 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into the mechanism of biological methanol activation based on the crystal structure of the methanol-cobalamin methyltransferase complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
8G3H
 
 | |
7XCN
 
 | | Crystal structure of the MttB-MttC complex at 2.7 A resolution | | Descriptor: | 5-HYDROXYBENZIMIDAZOLYLCOBAMIDE, GLYCEROL, Trimethylamine methyltransferase, ... | | Authors: | Li, J, Chan, M.K. | | Deposit date: | 2022-03-24 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into pyrrolysine function from structures of a trimethylamine methyltransferase and its corrinoid protein complex.
Commun Biol, 6, 2023
|
|
4JGI
 
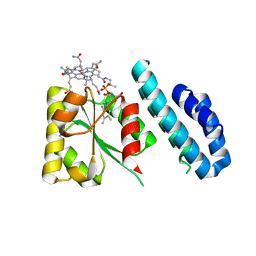 | | 1.5 Angstrom crystal structure of a novel cobalamin-binding protein from Desulfitobacterium hafniense DCB-2 | | Descriptor: | CO-METHYLCOBALAMIN, Putative uncharacterized protein | | Authors: | Sjuts, H, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2013-03-01 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the cobalamin-binding protein of a putative O-demethylase from Desulfitobacterium hafniense DCB-2.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1Y80
 
 | | Structure of a corrinoid (factor IIIm)-binding protein from Moorella thermoacetica | | Descriptor: | CO-5-METHOXYBENZIMIDAZOLYLCOBAMIDE, Predicted cobalamin binding protein, UNKNOWN ATOM OR ION | | Authors: | Liu, Z.-J, Fu, Z.-Q, Tempel, W, Das, A, Habel, J, Zhou, W, Chang, J, Chen, L, Lee, D, Nguyen, D, Chang, S.-H, Ljungdahl, L, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a corrinoid (factor IIIm)-binding protein from Moorella thermoacetica
To be published
|
|
3BUL
 
 | |
3IV9
 
 | |
1K7Y
 
 | | E. coli MetH C-terminal fragment (649-1227) | | Descriptor: | COBALAMIN, SULFATE ION, methionine synthase | | Authors: | Bandarian, V, Pattridge, K.A, Lennon, B.W, Huddler, D.P, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2001-10-22 | | Release date: | 2001-12-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Domain alternation switches B(12)-dependent methionine synthase to the activation conformation.
Nat.Struct.Biol., 9, 2002
|
|
3IVA
 
 | | Structure of the B12-dependent Methionine Synthase (MetH) C-teminal half with AdoHcy bound | | Descriptor: | COBALAMIN, Methionine synthase, NITRATE ION, ... | | Authors: | Pattridge, K.A, Koutmos, M, Smith, J.L. | | Deposit date: | 2009-08-31 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the reactivation of cobalamin-dependent methionine synthase.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1K98
 
 | | AdoMet complex of MetH C-terminal fragment | | Descriptor: | COBALAMIN, Methionine synthase, SULFATE ION | | Authors: | Bandarian, V, Pattridge, K.A, Lennon, B.W, Huddler, D.P, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2001-10-27 | | Release date: | 2001-12-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Domain alternation switches B(12)-dependent methionine synthase to the activation conformation.
Nat.Struct.Biol., 9, 2002
|
|
