2BTI
 
 | | Structure-function studies of the RmsA CsrA post-transcriptional global regulator protein family reveals a class of RNA-binding structure | | Descriptor: | ACETATE ION, CARBON STORAGE REGULATOR HOMOLOG, SULFATE ION | | Authors: | Heeb, S, Kuehne, S.A, Bycroft, M, Crivii, S, Allen, M.D, Haas, D, Camara, M, Williams, P. | | Deposit date: | 2005-05-31 | | Release date: | 2006-01-04 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Analysis of the Post-Transcriptional Regulator Rsma Reveals a Novel RNA-Binding Site.
J.Mol.Biol., 355, 2006
|
|
2JPP
 
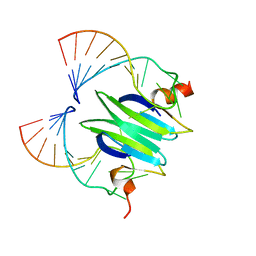 | | Structural basis of RsmA/CsrA RNA recognition: Structure of RsmE bound to the Shine-Dalgarno sequence of hcnA mRNA | | Descriptor: | RNA (5'-R(*GP*GP*GP*CP*UP*UP*CP*AP*CP*GP*GP*AP*UP*GP*AP*AP*GP*CP*CP*C)-3'), Translational repressor | | Authors: | Schubert, M, Lapouge, K, Duss, O, Oberstrass, F.C, Jelesarov, I, Haas, D, Allain, F.H.-T. | | Deposit date: | 2007-05-21 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA
Nat.Struct.Mol.Biol., 14, 2007
|
|
7YR6
 
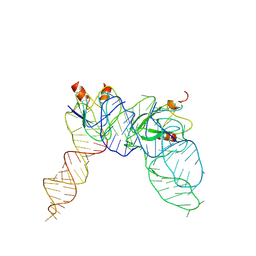 | | Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with two RsmA protein dimers | | Descriptor: | RsmZ RNA, Translational regulator CsrA | | Authors: | Jia, X, Pan, Z, Yuan, Y, Luo, B, Luo, Y, Mukherjee, S, Jia, G, Ling, X, Yang, X, Wu, Y, Liu, T, Wei, X, Bujnick, J.M, Zhao, K, Su, Z. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of sRNA RsmZ regulation of Pseudomonas aeruginosa virulence.
Cell Res., 33, 2023
|
|
7YR7
 
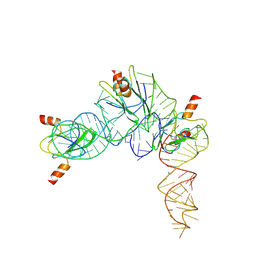 | | Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with three RsmA protein dimers | | Descriptor: | RsmZ RNA (118-MER), Translational regulator CsrA | | Authors: | Jia, X, Pan, Z, Yuan, Y, Luo, B, Luo, Y, Mukherjee, S, Jia, G, Liu, L, Ling, X, Yang, X, Wu, Y, Liu, T, Miao, Z, Wei, X, Bujnicki, J.M, Zhao, K, Su, Z. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of sRNA RsmZ regulation of Pseudomonas aeruginosa virulence.
Cell Res., 33, 2023
|
|
4K59
 
 | |
4KJI
 
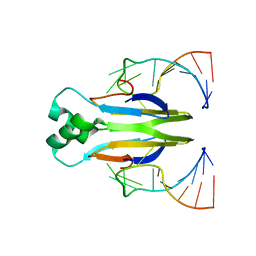 | |
4KRW
 
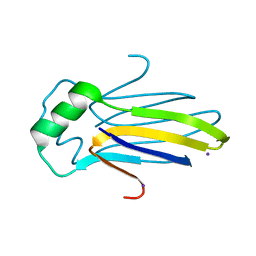 | |
1T3O
 
 | | Solution structure of CsrA, a bacterial carbon storage regulatory protein | | Descriptor: | Carbon storage regulator | | Authors: | Koharudin, L.M.I, Georgiou, T, Kleanthous, C, Geoffrey, R, Kaptein, R, Boelens, R. | | Deposit date: | 2004-04-27 | | Release date: | 2005-10-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | A model for RNA binding by the bacterial carbon storage regulatory protein, CsrA
To be Published
|
|
1VPZ
 
 | |
1Y00
 
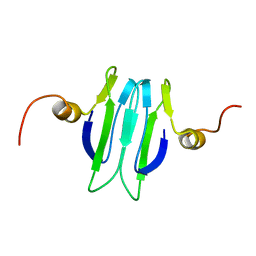 | | Solution structure of the Carbon Storage Regulator protein CsrA | | Descriptor: | Carbon storage regulator | | Authors: | Gutierrez, P, Li, Y, Osborne, M.J, Liu, Q, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-11-13 | | Release date: | 2005-06-21 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the carbon storage regulator protein CsrA from Escherichia coli.
J.Bacteriol., 187, 2005
|
|
2MFF
 
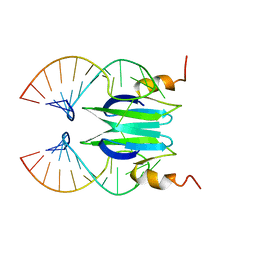 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL3)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL3(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2014-05-14 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFH
 
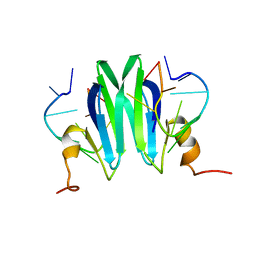 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(36-44)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, RsmZ(36-44) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2014-05-14 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFE
 
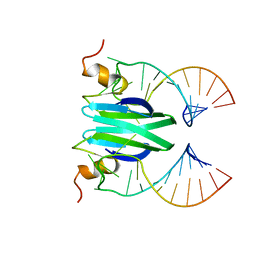 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL2)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL2(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2014-05-14 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFG
 
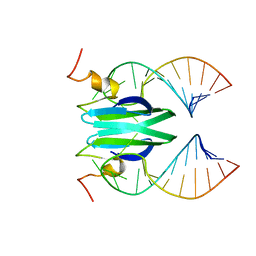 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL4)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL4(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2014-05-14 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFC
 
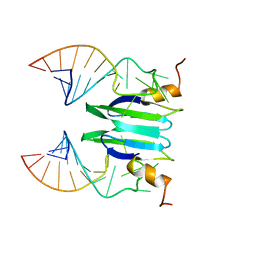 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL1)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL1(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-10 | | Release date: | 2014-02-26 | | Last modified: | 2014-05-14 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MF0
 
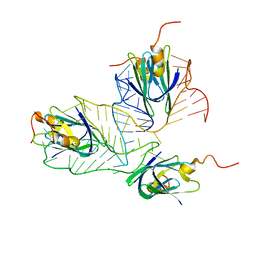 | | Structural basis of the non-coding RNA RsmZ acting as protein sponge: Conformer L of RsmZ(1-72)/RsmE(dimer) 1to3 complex | | Descriptor: | Carbon storage regulator homolog, RNA_(72-MER) | | Authors: | Duss, O, Michel, E, Yulikov, M, Schubert, M, Jeschke, G, Allain, F.H.-T. | | Deposit date: | 2013-10-02 | | Release date: | 2014-05-21 | | Last modified: | 2014-07-02 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the non-coding RNA RsmZ acting as a protein sponge.
Nature, 509, 2014
|
|
2MF1
 
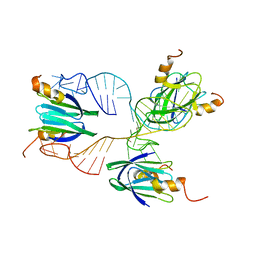 | | Structural basis of the non-coding RNA RsmZ acting as protein sponge: Conformer R of RsmZ(1-72)/RsmE(dimer) 1to3 complex | | Descriptor: | Carbon storage regulator homolog, RNA_(72-MER) | | Authors: | Duss, O, Michel, E, Yulikov, M, Schubert, M, Jeschke, G, Allain, F.H.-T. | | Deposit date: | 2013-10-02 | | Release date: | 2014-05-21 | | Last modified: | 2014-07-02 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the non-coding RNA RsmZ acting as a protein sponge.
Nature, 509, 2014
|
|
5Z38
 
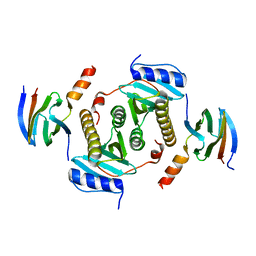 | | Crystal structure of CsrA bound to CesT | | Descriptor: | CesT protein, Truncated-CsrA, wild type CsrA | | Authors: | Ye, F, Yang, F, Yu, R, Lin, X, Qi, J, Chen, Z, Gao, G.F, Lu, G. | | Deposit date: | 2018-01-05 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Molecular basis of binding between the global post-transcriptional regulator CsrA and the T3SS chaperone CesT
Nat Commun, 9, 2018
|
|
5DMB
 
 | |
