1F48
 
 | | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI ARSENITE-TRANSLOCATING ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ANTIMONY (III) ION, ARSENITE-TRANSLOCATING ATPASE, ... | | Authors: | Zhou, T, Radaev, S, Rosen, B.P, Gatti, D.L. | | Deposit date: | 2000-06-07 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the ArsA ATPase: the catalytic subunit of a heavy metal resistance pump.
EMBO J., 19, 2000
|
|
3ZS9
 
 | | S. cerevisiae Get3-ADP-AlF4- complex with a cytosolic Get2 fragment | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPASE GET3, GOLGI TO ER TRAFFIC PROTEIN 2, ... | | Authors: | Mariappan, M, Mateja, A, Dobosz, M, Bove, E, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2011-06-24 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | The Mechanism of Membrane-Associated Steps in Tail-Anchored Protein Insertion.
Nature, 477, 2011
|
|
2WOO
 
 | | Nucleotide-free form of S. pombe Get3 | | Descriptor: | ATPASE GET3, ZINC ION | | Authors: | Mateja, A, Szlachcic, A, Downing, M.E, Dobosz, M, Mariappan, M, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2009-07-27 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.006 Å) | | Cite: | The Structural Basis of Tail-Anchored Membrane Protein Recognition by Get3.
Nature, 461, 2009
|
|
2WOJ
 
 | | ADP-AlF4 complex of S. cerevisiae GET3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPASE GET3, MAGNESIUM ION, ... | | Authors: | Mateja, A, Szlachcic, A, Downing, M.E, Dobosz, M, Mariappan, M, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2009-07-26 | | Release date: | 2009-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | The Structural Basis of Tail-Anchored Membrane Protein Recognition by Get3.
Nature, 461, 2009
|
|
3A36
 
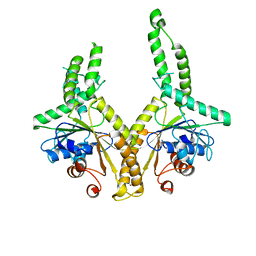 | | Structural insight into the membrane insertion of tail-anchored proteins by Get3 | | Descriptor: | ATPase GET3, ZINC ION | | Authors: | Yamagata, A, Mimura, H, Sato, Y, Yamashita, M, Yoshikawa, A, Fukai, S. | | Deposit date: | 2009-06-10 | | Release date: | 2010-01-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insight into the membrane insertion of tail-anchored proteins by Get3
Genes Cells, 15, 2010
|
|
3A37
 
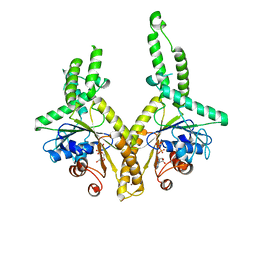 | | Structural insight into the membrane insertion of tail-anchored proteins by Get3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase GET3, ZINC ION | | Authors: | Yamagata, A, Mimura, H, Sato, Y, Yamashita, M, Yoshikawa, A, Fukai, S. | | Deposit date: | 2009-06-10 | | Release date: | 2010-01-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insight into the membrane insertion of tail-anchored proteins by Get3
Genes Cells, 15, 2010
|
|
4XTR
 
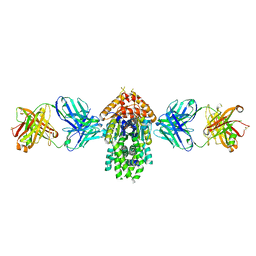 | | Structure of Get3 bound to the transmembrane domain of Pep12 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-23 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
4XVU
 
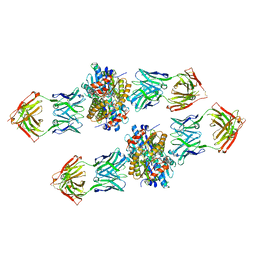 | | Structure of Get3 bound to the transmembrane domain of Nyv1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, Antibody heavy chain, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-28 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
4XWO
 
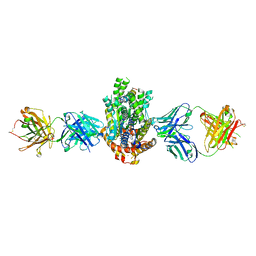 | | Structure of Get3 bound to the transmembrane domain of Sec22 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-29 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
1IHU
 
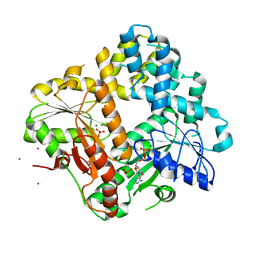 | | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI ARSENITE-TRANSLOCATING ATPASE IN COMPLEX WITH MG-ADP-ALF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ARSENICAL PUMP-DRIVING ATPASE, ... | | Authors: | Zhou, T, Radaev, S, Rosen, B.P, Gatti, D.L. | | Deposit date: | 2001-04-20 | | Release date: | 2001-09-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational changes in four regions of the Escherichia coli ArsA ATPase link ATP hydrolysis to ion translocation.
J.Biol.Chem., 276, 2001
|
|
1II9
 
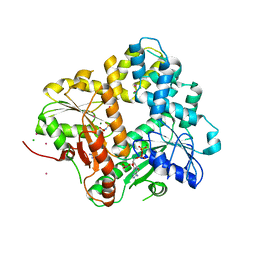 | | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI ARSENITE-TRANSLOCATING ATPASE IN COMPLEX WITH AMP-PNP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARSENICAL PUMP-DRIVING ATPASE, CADMIUM ION, ... | | Authors: | Zhou, T, Radaev, S, Gatti, D.L, Rosen, B.P. | | Deposit date: | 2001-04-21 | | Release date: | 2001-09-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational changes in four regions of the Escherichia coli ArsA ATPase link ATP hydrolysis to ion translocation.
J.Biol.Chem., 276, 2001
|
|
1II0
 
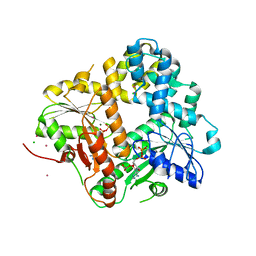 | | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI ARSENITE-TRANSLOCATING ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARSENICAL PUMP-DRIVING ATPASE, ... | | Authors: | Zhou, T, Radaev, S, Rosen, B.P, Gatti, D.L. | | Deposit date: | 2001-04-20 | | Release date: | 2001-09-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational changes in four regions of the Escherichia coli ArsA ATPase link ATP hydrolysis to ion translocation.
J.Biol.Chem., 276, 2001
|
|
5ZME
 
 | |
5ZMF
 
 | | AMPPNP complex of C. reinhardtii ArsA1 | | Descriptor: | ATPase ARSA1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Lin, T.W, Hsiao, C.D, Chang, H.Y. | | Deposit date: | 2018-04-03 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.556 Å) | | Cite: | Structural analysis of chloroplast tail-anchored membrane protein recognition by ArsA1.
Plant J., 99, 2019
|
|
6BS3
 
 | | Crystal structure of ADP-bound bacterial Get3-like A and B in Mycobacterium tuberculosis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Anion transporter, CALCIUM ION, ... | | Authors: | Li, H, Hu, K, Kovach, A. | | Deposit date: | 2017-12-01 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of Guided Entry of Tail-Anchored Proteins 3 Homologues in Mycobacterium tuberculosis.
J.Bacteriol., 201, 2019
|
|
6BS5
 
 | | Crystal structure of AMP-PNP-bound bacterial Get3-like A and B in Mycobacterium tuberculosis | | Descriptor: | Anion transporter, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, H, Hu, K, Kovach, A. | | Deposit date: | 2017-12-01 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Characterization of Guided Entry of Tail-Anchored Proteins 3 Homologues in Mycobacterium tuberculosis.
J.Bacteriol., 201, 2019
|
|
6BS4
 
 | | Crystal structure of ATPgammaS-bound bacterial Get3-like A and B in Mycobacterium tuberculosis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Anion transporter, MAGNESIUM ION, ... | | Authors: | Li, H, Hu, K, Kovach, A. | | Deposit date: | 2017-12-01 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of Guided Entry of Tail-Anchored Proteins 3 Homologues in Mycobacterium tuberculosis.
J.Bacteriol., 201, 2019
|
|
7SPY
 
 | | Get3 bound to ATP from G. intestinalis in the closed form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase ASNA1 homolog, MAGNESIUM ION, ... | | Authors: | Fry, M.Y, Maggiolo, A.O, Clemons Jr, W.M. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structurally derived universal mechanism for the catalytic cycle of the tail-anchored targeting factor Get3.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SPZ
 
 | | Nucleotide-free Get3 in two open forms | | Descriptor: | ATPase ASNA1 homolog, ZINC ION | | Authors: | Fry, M.Y, Maggiolo, A.O, Clemons Jr, W.M. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structurally derived universal mechanism for the catalytic cycle of the tail-anchored targeting factor Get3.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3H84
 
 | | Crystal structure of GET3 | | Descriptor: | ATPase GET3, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Hu, J, Li, J, Qian, X, Sha, B. | | Deposit date: | 2009-04-28 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structures of yeast Get3 suggest a mechanism for tail-anchored protein membrane insertion.
Plos One, 4, 2009
|
|
3IDQ
 
 | | Crystal structure of S. cerevisiae Get3 at 3.7 Angstrom resolution | | Descriptor: | ATPase GET3, NICKEL (II) ION, ZINC ION | | Authors: | Suloway, C.J.M, Chartron, J.W, Zaslaver, M, Clemons Jr, W.M. | | Deposit date: | 2009-07-21 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.701 Å) | | Cite: | Model for eukaryotic tail-anchored protein binding based on the structure of Get3
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3IQX
 
 | | ADP complex of C.therm. Get3 in closed form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Tail-anchored protein targeting factor Get3, ... | | Authors: | Bozkurt, G, Wild, K, Sinning, I. | | Deposit date: | 2009-08-21 | | Release date: | 2009-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insights into tail-anchored protein binding and membrane insertion by Get3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3IBG
 
 | | Crystal structure of Aspergillus fumigatus Get3 with bound ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase, subunit of the Get complex | | Authors: | Suloway, C.J.M, Chartron, J.W, Zaslaver, M, Clemons Jr, W.M. | | Deposit date: | 2009-07-15 | | Release date: | 2009-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Model for eukaryotic tail-anchored protein binding based on the structure of Get3
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3IO3
 
 | | GEt3 with ADP from D. Hansenii in Closed form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DEHA2D07832p, GLYCEROL, ... | | Authors: | Hu, J, Li, J, Qian, X, Sha, B. | | Deposit date: | 2009-08-13 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of yeast Get3 suggest a mechanism for tail-anchored protein membrane insertion
Plos One, 4, 2009
|
|
3IQW
 
 | | AMPPNP complex of C. therm. Get3 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Tail-anchored protein targeting factor Get3, ... | | Authors: | Bozkurt, G, Wild, K, Sinning, I. | | Deposit date: | 2009-08-21 | | Release date: | 2009-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into tail-anchored protein binding and membrane insertion by Get3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
