1DUJ
 
 | | SOLUTION STRUCTURE OF THE SPINDLE ASSEMBLY CHECKPOINT PROTEIN HUMAN MAD2 | | Descriptor: | SPINDLE ASSEMBLY CHECKPOINT PROTEIN | | Authors: | Luo, X, Fang, G, Coldiron, M, Lin, Y, Yu, H. | | Deposit date: | 2000-01-17 | | Release date: | 2000-03-08 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20.
Nat.Struct.Biol., 7, 2000
|
|
1GO4
 
 | | Crystal structure of Mad1-Mad2 reveals a conserved Mad2 binding motif in Mad1 and Cdc20. | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD1, Mitotic spindle assembly checkpoint protein MAD2A | | Authors: | Sironi, L, Mapelli, M, Jeang, K.T, Musacchio, A. | | Deposit date: | 2001-10-17 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a 'safety belt' binding mechanism for the spindle checkpoint.
EMBO J., 21, 2002
|
|
1KLQ
 
 | | The Mad2 Spindle Checkpoint Protein Undergoes Similar Major Conformational Changes upon Binding to Either Mad1 or Cdc20 | | Descriptor: | MITOTIC SPINDLE ASSEMBLY CHECKPOINT PROTEIN MAD2A, Mad2-binding peptide | | Authors: | Luo, X, Tang, Z, Rizo, J, Yu, H. | | Deposit date: | 2001-12-12 | | Release date: | 2002-01-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20.
Mol.Cell, 9, 2002
|
|
1S2H
 
 | | The Mad2 spindle checkpoint protein possesses two distinct natively folded states | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2A | | Authors: | Luo, X, Tang, Z, Xia, G, Wassmann, K, Matsumoto, T, Rizo, J, Yu, H. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-30 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | The Mad2 spindle checkpoint protein has two distinct natively folded states.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2QYF
 
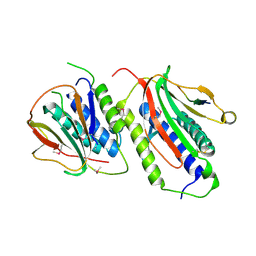 | |
2V64
 
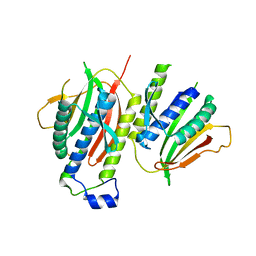 | | Crystallographic structure of the conformational dimer of the Spindle Assembly Checkpoint protein Mad2. | | Descriptor: | MBP1, MITOTIC SPINDLE ASSEMBLY CHECKPOINT PROTEIN MAD2A | | Authors: | Mapelli, M, Massimiliano, L, Santaguida, S, Musacchio, A. | | Deposit date: | 2007-07-13 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The MAD2 Conformational Dimer: Structure and Implications for the Spindle Assembly Checkpoint
Cell(Cambridge,Mass.), 131, 2007
|
|
2VFX
 
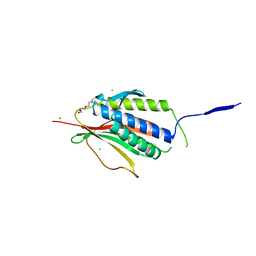 | | Structure of the Symmetric Mad2 Dimer | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, ... | | Authors: | Yang, M, Li, B, Liu, C.-J, Tomchick, D.R, Machius, M, Rizo, J, Yu, H, Luo, X. | | Deposit date: | 2007-11-05 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights Into MAD2 Regulation in the Spindle Checkpoint Revealed by the Crystal Structure of the Symmetric MAD2 Dimer.
Plos Biol., 6, 2008
|
|
3ABD
 
 | | Structure of human REV7 in complex with a human REV3 fragment in a monoclinic crystal | | Descriptor: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Hara, K, Hashimoto, H, Murakumo, Y, Kobayashi, S, Kogame, T, Unzai, S, Akashi, S, Takeda, S, Shimizu, T, Sato, M. | | Deposit date: | 2009-12-07 | | Release date: | 2010-02-16 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase {zeta} and REV1
J.Biol.Chem., 285, 2010
|
|
3ABE
 
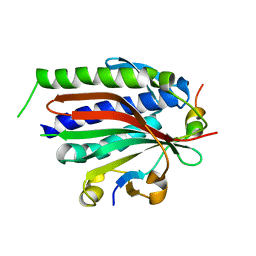 | | Structure of human REV7 in complex with a human REV3 fragment in a tetragonal crystal | | Descriptor: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Hara, K, Hashimoto, H, Murakumo, Y, Kobayashi, S, Kogame, T, Unzai, S, Akashi, S, Takeda, S, Shimizu, T, Sato, M. | | Deposit date: | 2009-12-07 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase {zeta} and REV1
J.Biol.Chem., 285, 2010
|
|
3GMH
 
 | | Crystal Structure of the Mad2 Dimer | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2A, SULFATE ION | | Authors: | Ozkan, E, Luo, X, Machius, M, Yu, H, Deisenhofer, J. | | Deposit date: | 2009-03-13 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structure of an intermediate conformer of the spindle checkpoint protein Mad2.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3VU7
 
 | | Crystal structure of REV1-REV7-REV3 ternary complex | | Descriptor: | DNA polymerase zeta catalytic subunit, DNA repair protein REV1, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Kikuchi, S, Hara, K, Shimizu, T, Sato, M, Hashimoto, H. | | Deposit date: | 2012-06-20 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of recruitment of DNA polymerase [zeta] by interaction between REV1 and REV7 proteins
J.Biol.Chem., 287, 2012
|
|
4AEZ
 
 | | Crystal Structure of Mitotic Checkpoint Complex | | Descriptor: | MITOTIC SPINDLE CHECKPOINT COMPONENT MAD2, MITOTIC SPINDLE CHECKPOINT COMPONENT MAD3, WD REPEAT-CONTAINING PROTEIN SLP1 | | Authors: | Kulkarni, K.A, Chao, W.C.H, Zhang, Z, Barford, D. | | Deposit date: | 2012-01-13 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Mitotic Checkpoint Complex
Nature, 484, 2012
|
|
4EXT
 
 | |
4FJO
 
 | | Structure of the Rev1 CTD-Rev3/7-Pol kappa RIR complex | | Descriptor: | DNA polymerase kappa, DNA polymerase zeta catalytic subunit, DNA repair protein REV1, ... | | Authors: | Wojtaszek, J, Lee, C.-J, Zhou, P. | | Deposit date: | 2012-06-11 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.718 Å) | | Cite: | Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric Pol zeta and Pol kappa
J.Biol.Chem., 287, 2012
|
|
4GK0
 
 | | Crystal structure of human Rev3-Rev7-Rev1 complex | | Descriptor: | DNA polymerase zeta catalytic subunit, DNA repair protein REV1, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Tao, J, Min, X, Wei, X. | | Deposit date: | 2012-08-10 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the assembly of human translesion polymerase complexes
Protein Cell, 3, 2012
|
|
4GK5
 
 | | Crystal structure of human Rev3-Rev7-Rev1-Polkappa complex | | Descriptor: | DNA polymerase kappa, DNA polymerase zeta catalytic subunit, DNA repair protein REV1, ... | | Authors: | Tao, J, Min, X, Wei, X. | | Deposit date: | 2012-08-10 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural insights into the assembly of human translesion polymerase complexes
Protein Cell, 3, 2012
|
|
4TRK
 
 | | Structure of C. elegans HIM-3 | | Descriptor: | C. elegans HIM-3 | | Authors: | Rosenberg, S.C, Corbett, K.D. | | Deposit date: | 2014-06-17 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The Chromosome Axis Controls Meiotic Events through a Hierarchical Assembly of HORMA Domain Proteins.
Dev.Cell, 31, 2014
|
|
4TZJ
 
 | |
4TZL
 
 | |
4TZM
 
 | |
4TZN
 
 | | Structure of HTP-2 bound to HTP-3 motif-6 | | Descriptor: | Protein HTP-2, Protein HTP-3 | | Authors: | Rosenberg, S.C, Corbett, K.D. | | Deposit date: | 2014-07-10 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.115 Å) | | Cite: | The Chromosome Axis Controls Meiotic Events through a Hierarchical Assembly of HORMA Domain Proteins.
Dev.Cell, 31, 2014
|
|
4TZO
 
 | |
4TZQ
 
 | |
4TZS
 
 | |
5KHU
 
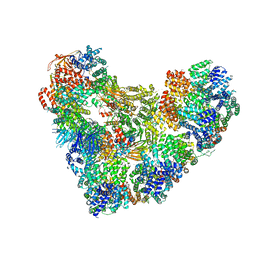 | | Model of human Anaphase-promoting complex/Cyclosome (APC15 deletion mutant), in complex with the Mitotic checkpoint complex (APC/C-CDC20-MCC) based on cryo EM data at 4.8 Angstrom resolution | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Yamaguchi, M, VanderLinden, R, Dube, P, Stark, H, Schulman, B. | | Deposit date: | 2016-06-15 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM of Mitotic Checkpoint Complex-Bound APC/C Reveals Reciprocal and Conformational Regulation of Ubiquitin Ligation.
Mol.Cell, 63, 2016
|
|
