4M51
 
 | | Crystal structure of amidohydrolase nis_0429 (ser145ala mutant) from nitratiruptor sp. sb155-2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Amidohydrolase family protein, BENZOIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Gobble, A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-07 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Deamination of 6-aminodeoxyfutalosine in menaquinone biosynthesis by distantly related enzymes.
Biochemistry, 52, 2013
|
|
6HFQ
 
 | |
6SJ3
 
 | | Amidohydrolase, AHS with 3-HBA | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXYBENZOIC ACID, Amidohydrolase, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6HFP
 
 | |
6HFJ
 
 | |
5LXX
 
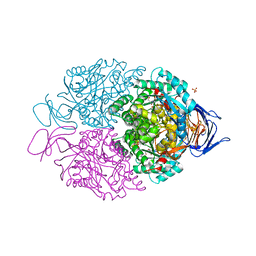 | | High-resolution structure of human collapsin response mediator protein 2 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydropyrimidinase-related protein 2, SULFATE ION | | Authors: | Myllykoski, M, Hensley, K, Kursula, P. | | Deposit date: | 2016-09-23 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Collapsin response mediator protein 2: high-resolution crystal structure sheds light on small-molecule binding, post-translational modifications, and conformational flexibility.
Amino Acids, 49, 2017
|
|
6SJ2
 
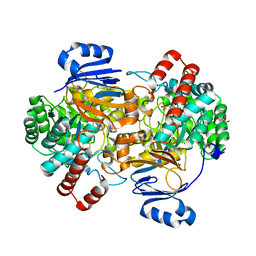 | | Amidohydrolase, AHS with 3-HAA | | Descriptor: | 3-HYDROXYANTHRANILIC ACID, Amidohydrolase, GLYCEROL, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2020-04-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4C6F
 
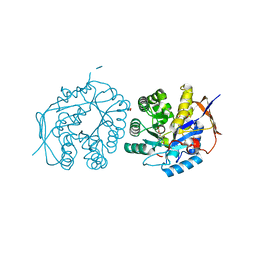 | | Crystal structure of the dihydroorotase domain of human CAD bound to substrate at pH 6.5 | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, DIHYDROOROTASE, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.261 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
4C6E
 
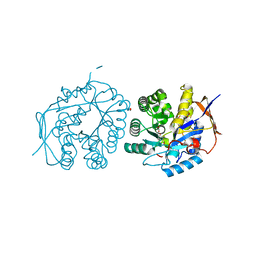 | | Crystal structure of the dihydroorotase domain of human CAD bound to substrate at pH 5.5 | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, DIHYDROOROTASE, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.263 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
5OL4
 
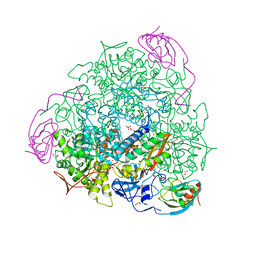 | | 1.28 A resolution of Sporosarcina pasteurii urease inhibited in the presence of NBPT | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Phosphoramidothioic O,O-acid, ... | | Authors: | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | Deposit date: | 2017-07-26 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Urease Inhibition in the Presence of N-(n-Butyl)thiophosphoric Triamide, a Suicide Substrate: Structure and Kinetics.
Biochemistry, 56, 2017
|
|
8PBM
 
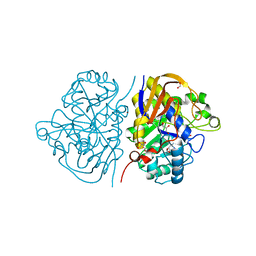 | |
2Z26
 
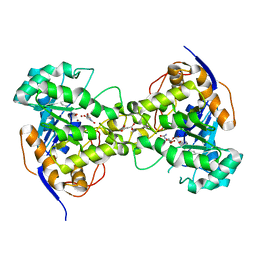 | | Thr110Ala dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, MALONATE ION, ... | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Kinetic and Structural Analysis of Mutant Escherichia coli Dihydroorotases: A Flexible Loop Stabilizes the Transition State
Biochemistry, 46, 2007
|
|
1GKP
 
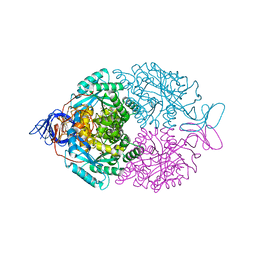 | | D-Hydantoinase (Dihydropyrimidinase) from Thermus sp. in space group C2221 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HYDANTOINASE, SULFATE ION, ... | | Authors: | Abendroth, J, Niefind, K, Schomburg, D. | | Deposit date: | 2001-08-20 | | Release date: | 2002-06-27 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.295 Å) | | Cite: | X-Ray Structure of a Dihydropyrimidinase from Thermus Sp. At 1.3 A Resolution
J.Mol.Biol., 320, 2002
|
|
4C6D
 
 | | Crystal structure of the dihydroorotase domain of human CAD bound to substrate at pH 6.0 | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CAD PROTEIN, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
4C6J
 
 | | Crystal structure of the dihydroorotase domain of human CAD bound to substrate at pH 7.5 | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CAD PROTEIN, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
6HFR
 
 | |
6RKG
 
 | | 1.32 A RESOLUTION OF SPOROSARCINA PASTEURII UREASE INHIBITED IN THE PRESENCE OF NBPTO AT pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, DIAMIDOPHOSPHATE, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Benini, S, Ciurli, S. | | Deposit date: | 2019-04-30 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The Impact of pH on Catalytically Critical Protein Conformational Changes: The Case of the Urease, a Nickel Enzyme.
Chemistry, 25, 2019
|
|
6HFS
 
 | |
4C6I
 
 | | Crystal structure of the dihydroorotase domain of human CAD bound to substrate at pH 7.0 | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CAD PROTEIN, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
3V7P
 
 | | Crystal structure of amidohydrolase nis_0429 (target efi-500396) from Nitratiruptor sp. sb155-2 | | Descriptor: | Amidohydrolase family protein, BENZOIC ACID, BICARBONATE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-12-21 | | Release date: | 2012-01-11 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Amidohydrolase Nis_0429 (Target Efi-500319) from Nitratiruptor Sp. Sb155-2
To be Published
|
|
6HFL
 
 | |
6HFU
 
 | |
3LS9
 
 | | Crystal structure of atrazine chlorohydrolase TrzN from Arthrobacter aurescens TC1 complexed with zinc | | Descriptor: | Triazine hydrolase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Seffernick, J, Wackett, L.P, Almo, S.C. | | Deposit date: | 2010-02-12 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of atrazine chlorohydrolase TrzN
from Arthrobacter aurescens TC1 complexed with zinc
To be Published
|
|
3MDU
 
 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-Guanidino-L-Glutamate | | Descriptor: | GLYCEROL, N-carbamimidoyl-L-glutamic acid, N-formimino-L-Glutamate Iminohydrolase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-03-30 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4003 Å) | | Cite: | Structure of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa.
Biochemistry, 54, 2015
|
|
6QDY
 
 | | The crystal structure of Sporosarcina pasteurii urease in complex with its substrate urea | | Descriptor: | 1,2-ETHANEDIOL, FLUORIDE ION, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Benini, S, Ciurli, S. | | Deposit date: | 2019-01-03 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.416 Å) | | Cite: | The Structure of the Elusive Urease-Urea Complex Unveils the Mechanism of a Paradigmatic Nickel-Dependent Enzyme.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
