2HEK
 
 | | Crystal structure of O67745, a hypothetical protein from Aquifex aeolicus at 2.0 A resolution. | | Descriptor: | BROMIDE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Oganesyan, V, Jancarik, J, Adams, P.D, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structure of O67745_AQUAE, a hypothetical protein from Aquifex aeolicus.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
3NQW
 
 | | A metazoan ortholog of SpoT hydrolyzes ppGpp and plays a role in starvation responses | | Descriptor: | CG11900, MANGANESE (II) ION, SULFATE ION | | Authors: | Sun, D.W, Kim, H.Y, Kim, K.J, Jeon, Y.H, Chung, J. | | Deposit date: | 2010-06-30 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses
Nat.Struct.Mol.Biol., 17, 2010
|
|
7S2Y
 
 | | SAMHD1 HD domain bound to CNDAC | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 4-amino-1-{2-cyano-2-deoxy-5-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-arabinofuranosyl}pyrimidin-2(1H)-one, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Digianantonio, K.M, Xiong, Y. | | Deposit date: | 2021-09-04 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Differences between intrinsic and acquired nucleoside analogue resistance in acute myeloid leukaemia cells.
J Exp Clin Cancer Res, 40, 2021
|
|
6TXF
 
 | | Crystal structure of tetrameric human D137N-SAMHD1 (residues 109-626) with XTP, dAMPNPP and Mn | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A.G, Kelly, G, Taylor, I.A. | | Deposit date: | 2020-01-14 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
7LTT
 
 | | SAMHD1(113-626) H206R D207N R366C | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, MAGNESIUM ION | | Authors: | Temple, J.T, Bowen, N.E. | | Deposit date: | 2021-02-20 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional characterization explains loss of dNTPase activity of the cancer-specific R366C/H mutant SAMHD1 proteins.
J.Biol.Chem., 297, 2021
|
|
7LU5
 
 | | SAMHD1(113-626) H206R D207N R366H | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1 | | Authors: | Temple, J.T, Bowen, N.E. | | Deposit date: | 2021-02-20 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.57 Å) | | Cite: | Structural and functional characterization explains loss of dNTPase activity of the cancer-specific R366C/H mutant SAMHD1 proteins.
J.Biol.Chem., 297, 2021
|
|
4X9E
 
 | | DEOXYGUANOSINETRIPHOSPHATE TRIPHOSPHOHYDROLASE from Escherichia coli with two DNA effector molecules | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, MAGNESIUM ION, RNA (5'-R(P*CP*CP*C)-3') | | Authors: | Singh, D, Gawel, D, Itsko, M, Krahn, J.M, London, R.E, Schaaper, R.M. | | Deposit date: | 2014-12-11 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Escherichia coli dGTP Triphosphohydrolase: A HEXAMERIC ENZYME WITH DNA EFFECTOR MOLECULES.
J.Biol.Chem., 290, 2015
|
|
3GW7
 
 | | Crystal structure of a metal-dependent phosphohydrolase with conserved HD domain (yedJ) from Escherichia coli in complex with nickel ions. Northeast Structural Genomics Consortium Target ER63 | | Descriptor: | NICKEL (II) ION, Uncharacterized protein yedJ | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Janjua, J, Xiao, R, Cunningham, K, Ma, L, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-31 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of a metal-dependent phosphohydrolase with conserved HD domain (yedJ) from Escherichia coli in complex with nickel ions. Northeast Structural Genomics Consortium Target ER63
To be Published
|
|
8GB2
 
 | | Crystal structure of Apo-SAMHD1 | | Descriptor: | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION | | Authors: | Egleston, M, Dong, L, Howlader, A.H, Bhat, S, Orris, B, Bianchet, M.A, Greenberg, M.M, Stivers, J.T. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Deoxyguanosine-Linked Bifunctional Inhibitor of SAMHD1 dNTPase Activity and Nucleic Acid Binding.
Acs Chem.Biol., 18, 2023
|
|
8GB1
 
 | | Crystal structure of SAMHD1 dimer bound to deoxyguanosine linked inhibitor | | Descriptor: | 5'-O-[(R)-(3-{[(1M)-3'-bromo[1,1'-biphenyl]-3-carbonyl]amino}propoxy)(hydroxy)phosphoryl]-2'-deoxyguanosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION | | Authors: | Egleston, M, Dong, L, Howlader, A.H, Bhat, S, Orris, B, Bianchet, M.A, Greenberg, M.M, Stivers, J.T. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Deoxyguanosine-Linked Bifunctional Inhibitor of SAMHD1 dNTPase Activity and Nucleic Acid Binding.
Acs Chem.Biol., 18, 2023
|
|
4BZC
 
 | | Crystal structure of the tetrameric dGTP-bound wild type SAMHD1 catalytic core | | Descriptor: | 2'-deoxyguanosine-5'-O-(1-thiotriphosphate), DEOXYNUCLEOSIDE TRIPHOSPHATE TRIPHOSPHOHYDROLASE SAMHD1, MAGNESIUM ION, ... | | Authors: | Ji, X, Yang, H, Wu, Y, Yan, J, Mehrens, J, DeLucia, M, Hao, C, Gronenborn, A.M, Skowronski, J, Ahn, J, Xiong, Y. | | Deposit date: | 2013-07-25 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Mechanism of Allosteric Activation of Samhd1 by Dgtp
Nat.Struct.Mol.Biol., 20, 2013
|
|
4BZB
 
 | | Crystal structure of the tetrameric dGTP-bound SAMHD1 mutant catalytic core | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DEOXYNUCLEOSIDE TRIPHOSPHATE TRIPHOSPHOHYDROLASE SAMHD1, MAGNESIUM ION | | Authors: | Ji, X, Yang, H, Wu, Y, Yan, J, Mehrens, J, DeLucia, M, Hao, C, Gronenborn, A.M, Skowronski, J, Ahn, J, Xiong, Y. | | Deposit date: | 2013-07-25 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Mechanism of Allosteric Activation of Samhd1 by Dgtp
Nat.Struct.Mol.Biol., 20, 2013
|
|
1YNB
 
 | | crystal structure of genomics APC5600 | | Descriptor: | hypothetical protein AF1432 | | Authors: | Dong, A, Skarina, T, Savchenko, A, Pai, E.F, Joachimiak, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-01-24 | | Release date: | 2005-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of genomics AF1432 by Sulfur SAD methods
To be Published
|
|
1YOY
 
 | |
8WMY
 
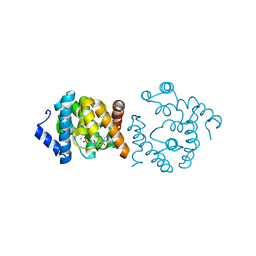 | | Crystal structure of YqeK in complex with MG and ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, bis(5'-nucleosyl)-tetraphosphatase (symmetrical) | | Authors: | Mo, X. | | Deposit date: | 2023-10-05 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular Mechanism of Ap4A Hydrolysis by YqeK of Staphylococcus pyogenes.
To be published
|
|
4XDS
 
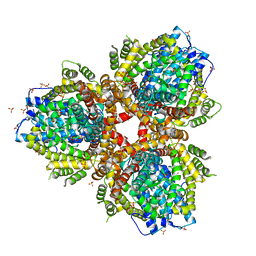 | | Deoxyguanosinetriphosphate Triphosphohydrolase from Escherichia coli with Nickel | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, NICKEL (II) ION, SULFATE ION | | Authors: | Singh, D, Gawel, D, Itsko, M, Krahn, J.M, London, R.E, Schaaper, R.M. | | Deposit date: | 2014-12-19 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.354 Å) | | Cite: | Structure of Escherichia coli dGTP Triphosphohydrolase: A HEXAMERIC ENZYME WITH DNA EFFECTOR MOLECULES.
J.Biol.Chem., 290, 2015
|
|
4ZWG
 
 | | Crystal structure of the GTP-dATP-bound catalytic core of SAMHD1 phosphomimetic T592E mutant | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Tang, C, Ji, X, Xiong, Y. | | Deposit date: | 2015-05-19 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Impaired dNTPase Activity of SAMHD1 by Phosphomimetic Mutation of Thr-592.
J.Biol.Chem., 290, 2015
|
|
4ZWE
 
 | |
2CQZ
 
 | |
5AO2
 
 | |
5AO4
 
 | |
5AO1
 
 | |
5AO3
 
 | |
3B57
 
 | | Crystal structure of the Lin1889 protein (Q92AN1) from Listeria innocua. Northeast Structural Consortium target LkR65 | | Descriptor: | Lin1889 protein, MAGNESIUM ION | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Benach, J, Forouhar, F, Vorobiev, S.M, Wang, H, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-25 | | Release date: | 2007-11-06 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Lin1889 protein (Q92AN1) from Listeria innocua.
To be Published
|
|
5TK7
 
 | |
