3K3T
 
 | | E185A mutant of peptidoglycan hydrolase from Sphingomonas sp. A1 | | Descriptor: | Peptidoglycan hydrolase FlgJ, SULFATE ION | | Authors: | Maruyama, Y, Ochiai, A, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-10-04 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mutational studies of the peptidoglycan hydrolase FlgJ of Sphingomonas sp. strain A1
J.Basic Microbiol., 50, 2010
|
|
3VWO
 
 | |
2ZYC
 
 | | Crystal structure of peptidoglycan hydrolase from Sphingomonas sp. A1 | | Descriptor: | PHOSPHATE ION, Peptidoglycan hydrolase FlgJ | | Authors: | Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-01-19 | | Release date: | 2009-02-03 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of the glycosidase family 73 peptidoglycan hydrolase FlgJ
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3FI7
 
 | | Crystal Structure of the autolysin Auto (Lmo1076) from Listeria monocytogenes, catalytic domain | | Descriptor: | Lmo1076 protein, SULFATE ION | | Authors: | Bublitz, M, Polle, L, Holland, C, Nimtz, M, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2008-12-11 | | Release date: | 2009-04-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for autoinhibition and activation of Auto, a virulence-associated peptidoglycan hydrolase of Listeria monocytogenes.
Mol.Microbiol., 71, 2009
|
|
4QDN
 
 | | Crystal Structure of the endo-beta-N-acetylglucosaminidase from Thermotoga maritima | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Flagellar protein FlgJ [peptidoglycan hydrolase], PHOSPHATE ION | | Authors: | Lipski, A, Nurizzo, D, Bourne, Y, Vincent, F. | | Deposit date: | 2014-05-14 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical characterization of the beta-N-acetylglucosaminidase from Thermotoga maritima: Toward rationalization of mechanistic knowledge in the GH73 family.
Glycobiology, 25, 2015
|
|
5DN4
 
 | |
5DN5
 
 | | Structure of a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ | | Descriptor: | CHLORIDE ION, IODIDE ION, Peptidoglycan hydrolase FlgJ, ... | | Authors: | Zaloba, P, Bailey-Elkin, B.A, Mark, B.L. | | Deposit date: | 2015-09-09 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Biochemical Insights into the Peptidoglycan Hydrolase Domain of FlgJ from Salmonella typhimurium.
Plos One, 11, 2016
|
|
6FXO
 
 | | Crystal structure of Major Bifunctional Autolysin | | Descriptor: | Bifunctional autolysin, CHLORIDE ION | | Authors: | Pintar, S, Turk, D. | | Deposit date: | 2018-03-09 | | Release date: | 2019-03-20 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Domain sliding of two Staphylococcus aureus N-acetylglucosaminidases enables their substrate-binding prior to its catalysis.
Commun Biol, 3, 2020
|
|
6FXP
 
 | | Crystal structure of S. aureus glucosaminidase B | | Descriptor: | CHLORIDE ION, SODIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Pintar, S, Turk, D. | | Deposit date: | 2018-03-09 | | Release date: | 2019-03-20 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Domain sliding of two Staphylococcus aureus N-acetylglucosaminidases enables their substrate-binding prior to its catalysis.
Commun Biol, 3, 2020
|
|
5WQW
 
 | | X-ray structure of catalytic domain of autolysin from Clostridium perfringens | | Descriptor: | 1,2-ETHANEDIOL, N-acetylglucosaminidase | | Authors: | Tamai, E, Sekiya, H, Goda, E, Makihata, N, Maki, J, Yoshida, H, Kamitori, S. | | Deposit date: | 2016-11-29 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and biochemical characterization of the Clostridium perfringens autolysin catalytic domain
FEBS Lett., 591, 2017
|
|
4PI9
 
 | | Crystal structure of S. Aureus Autolysin E in complex with muropeptide NAM-L-ALA-D-iGLU | | Descriptor: | (4R)-4-[[(2S)-2-[[(2R)-2-[(2R,3S,4R,5R,6R)-5-acetamido-2-(hydroxymethyl)-3,6-bis(oxidanyl)oxan-4-yl]oxypropanoyl]amino]propanoyl]amino]-5-azanyl-5-oxidanylidene-pentanoic acid, Autolysin E, CHLORIDE ION, ... | | Authors: | Mihelic, M, Renko, M, Turk, D. | | Deposit date: | 2014-05-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The mechanism behind the selection of two different cleavage sites in NAG-NAM polymers.
IUCrJ, 4, 2017
|
|
4PI7
 
 | | Crystal structure of S. Aureus Autolysin E in complex with disaccharide NAM-NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, Autolysin E, CHLORIDE ION, ... | | Authors: | Mihelic, M, Renko, M, Jakas, A, Turk, D. | | Deposit date: | 2014-05-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The mechanism behind the selection of two different cleavage sites in NAG-NAM polymers
Iucrj, 4, 2017
|
|
4PIA
 
 | | Crystal structure of S. Aureus Autolysin E | | Descriptor: | Autolysin E, CHLORIDE ION | | Authors: | Mihelic, M, Renko, M, Dobersek, A, Bedrac, L, Turk, D. | | Deposit date: | 2014-05-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.466 Å) | | Cite: | The mechanism behind the selection of two different cleavage sites in NAG-NAM polymers
Iucrj, 4, 2017
|
|
4PI8
 
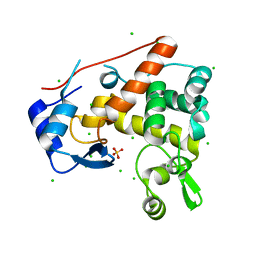 | | Crystal structure of catalytic mutant E138A of S. Aureus Autolysin E in complex with disaccharide NAG-NAM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, Autolysin E, CHLORIDE ION, ... | | Authors: | Mihelic, M, Renko, M, Jakas, A, Turk, D. | | Deposit date: | 2014-05-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | The mechanism behind the selection of two different cleavage sites in NAG-NAM polymers
Iucrj, 4, 2017
|
|
7QFU
 
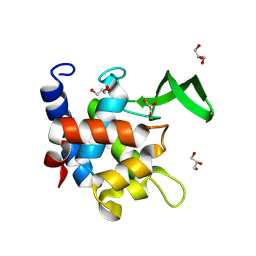 | | Crystal Structure of AtlA catalytic domain from Enterococcus feacalis | | Descriptor: | GLYCEROL, Peptidoglycan hydrolase | | Authors: | Zamboni, V, Barelier, S, Dixon, R, Galley, N, Ghanem, A, Cahuzac, H, Salamaga, B, Davis, P.J, Mesnage, S, Vincent, F. | | Deposit date: | 2021-12-06 | | Release date: | 2022-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular basis for substrate recognition and septum cleavage by AtlA, the major N-acetylglucosaminidase of Enterococcus faecalis.
J.Biol.Chem., 298, 2022
|
|
6U0O
 
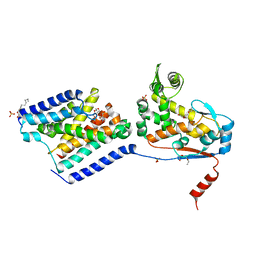 | | Crystal structure of a peptidoglycan release complex, SagB-SpdC, in lipidic cubic phase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(2-ETHOXYETHOXY)ETHANOL, CITRATE ANION, ... | | Authors: | Owens, T.W, Schaefer, K, Kahne, D, Walker, S. | | Deposit date: | 2019-08-14 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and reconstitution of a hydrolase complex that may release peptidoglycan from the membrane after polymerization.
Nat Microbiol, 6, 2021
|
|
4Q2W
 
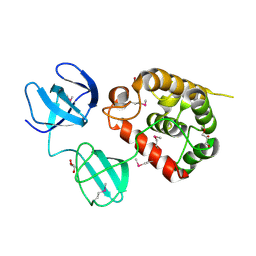 | | Crystal Structure of pneumococcal peptidoglycan hydrolase LytB | | Descriptor: | GLYCEROL, Putative endo-beta-N-acetylglucosaminidase | | Authors: | Bai, X.H, Chen, H.J, Jiang, Y.L, Wen, Z, Cheng, W, Li, Q, Zhang, J.R, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-16 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of pneumococcal peptidoglycan hydrolase LytB reveals insights into the bacterial cell wall remodeling and pathogenesis.
J.Biol.Chem., 289, 2014
|
|
5T1Q
 
 | | 2.15 Angstrom Crystal Structure of N-acetylmuramoyl-L-alanine Amidase from Staphylococcus aureus. | | Descriptor: | N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979, SODIUM ION, TRIETHYLENE GLYCOL | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bagnoli, F, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-19 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.15 Angstrom Crystal Structure of N-acetylmuramoyl-L-alanine Amidase from Staphylococcus aureus.
To Be Published
|
|
4KT3
 
 | | Structure of a type VI secretion system effector-immunity complex from Pseudomonas protegens | | Descriptor: | Putative lipoprotein, Uncharacterized protein | | Authors: | Whitney, J.C, Chou, S, Gardiner, T.E, Mougous, J.D. | | Deposit date: | 2013-05-19 | | Release date: | 2013-07-31 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (1.4362 Å) | | Cite: | Identification, Structure, and Function of a Novel Type VI Secretion Peptidoglycan Glycoside Hydrolase Effector-Immunity Pair.
J.Biol.Chem., 288, 2013
|
|
7PJ6
 
 | |
7PL3
 
 | |
7PJ4
 
 | |
7PJ3
 
 | |
7PJ5
 
 | |
7POD
 
 | |
