8B28
 
 | |
7XJP
 
 | | Cryo-EM structure of EDS1 and SAG101 with ATP-APDR | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5-DIPHOSPHORIBOSE, ISOPROPYL ALCOHOL, ... | | Authors: | Huang, S.J, Jia, A.L, Han, Z.F, Chai, J.J. | | Deposit date: | 2022-04-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | TIR-catalyzed ADP-ribosylation reactions produce signaling molecules for plant immunity.
Science, 377, 2022
|
|
7XEY
 
 | | EDS1-PAD4 complexed with pRib-ADP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-O-phosphono-beta-D-ribofuranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Huang, S, Jia, A, Xiao, Y. | | Deposit date: | 2022-03-31 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Identification and receptor mechanism of TIR-catalyzed small molecules in plant immunity.
Science, 377, 2022
|
|
7XDD
 
 | | Cryo-EM structure of EDS1 and PAD4 | | Descriptor: | Lipase-like PAD4, Protein EDS1 | | Authors: | Huang, S.J, Jia, A.L, Sun, Y, Han, Z.F, Chai, J.J. | | Deposit date: | 2022-03-26 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Identification and receptor mechanism of TIR-catalyzed small molecules in plant immunity.
Science, 377, 2022
|
|
7X0D
 
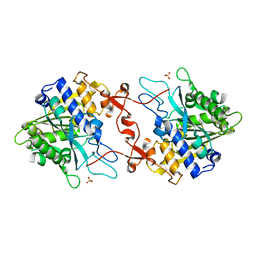 | | Crystal structure of phospholipase A1, CaPLA1 | | Descriptor: | Phospholipase A1, SULFATE ION | | Authors: | Heo, Y, Lee, I, Moon, S, Lee, W. | | Deposit date: | 2022-02-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39725161 Å) | | Cite: | Crystal Structures of the Plant Phospholipase A1 Proteins Reveal a Unique Dimerization Domain.
Molecules, 27, 2022
|
|
7X0C
 
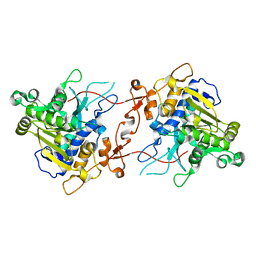 | | Crystal structure of phospholipase A1, AtDSEL | | Descriptor: | Phospholipase A1-IIgamma | | Authors: | Heo, Y, Lee, I, Moon, S, Lee, W. | | Deposit date: | 2022-02-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79909515 Å) | | Cite: | Crystal Structures of the Plant Phospholipase A1 Proteins Reveal a Unique Dimerization Domain.
Molecules, 27, 2022
|
|
7SPR
 
 | |
7APN
 
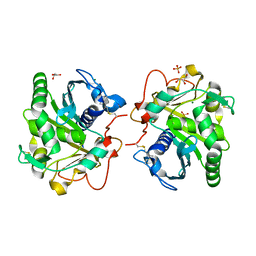 | | Structure of Lipase TL from bulk agarose grown crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Lipase, ... | | Authors: | Gavira, J.A, Martinez-Rodriguez, S, Fernande-Penas, R, Verdugo-Escamilla, C. | | Deposit date: | 2020-10-19 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Production of Cross-Linked Lipase Crystals at a Preparative Scale.
Cryst.Growth Des., 21, 2021
|
|
7APP
 
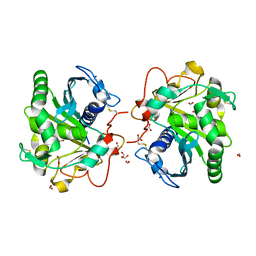 | | Structure of Lipase TL from capillary grown crystal in the presence of agarose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FORMIC ACID, Lipase, ... | | Authors: | Gavira, J.A, Martinez-Rodriguez, S, Fernande-Penas, R, Verdugo-Escamilla, C. | | Deposit date: | 2020-10-19 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Production of Cross-Linked Lipase Crystals at a Preparative Scale.
Cryst.Growth Des., 21, 2021
|
|
6XS3
 
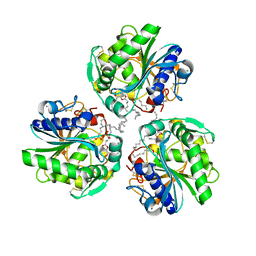 | |
6XRV
 
 | |
6XOK
 
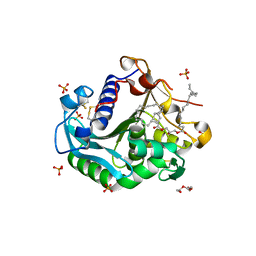 | |
6YXX
 
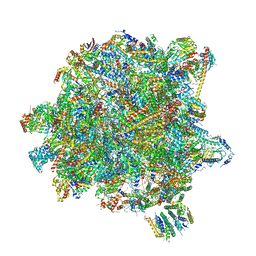 | | State A of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | Descriptor: | 12S ribosomal RNA, 50S ribosomal protein L13, putative, ... | | Authors: | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2020-05-04 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
6YXY
 
 | | State B of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | Descriptor: | 12S ribosomal RNA, ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2020-05-04 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
6L7N
 
 | |
6UNV
 
 | |
6OR3
 
 | |
6QPP
 
 | | Rhizomucor miehei lipase propeptide complex, native | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase | | Authors: | Moroz, O.V, Blagova, E, Reiser, V, Saikia, R, Dalal, S, Jorgensen, C.I, Baunsgaard, L, Andersen, B, Svendsen, A, Wilson, K.S. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Novel Inhibitory Function of theRhizomucor mieheiLipase Propeptide and Three-Dimensional Structures of Its Complexes with the Enzyme.
Acs Omega, 4, 2019
|
|
6QPR
 
 | | Rhizomucor miehei lipase propeptide complex, Ser95/Ile96 deletion mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase | | Authors: | Moroz, O.V, Blagova, E, Reiser, V, Saikia, R, Dalal, S, Jorgensen, C.I, Baunsgaard, L, Andersen, B, Svendsen, A, Wilson, K.S. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel Inhibitory Function of theRhizomucor mieheiLipase Propeptide and Three-Dimensional Structures of Its Complexes with the Enzyme.
Acs Omega, 4, 2019
|
|
6Q6Z
 
 | |
6I8G
 
 | |
6I8H
 
 | |
6HW1
 
 | | ROOM TEMPERATURE STRUCTURE OF LIPASE FROM T. LANUGINOSA AT 2.5 A RESOLUTION IN CHIPX MICROFLUIDIC DEVICE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase, MAGNESIUM ION, ... | | Authors: | Gavira, J.A, Fernadez-Penas, R, Martinez-Rodriguez, S, Verdugo-Escamilla, C. | | Deposit date: | 2018-10-11 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
6A0W
 
 | | Crystal structure of lipase from Rhizopus microsporus var. chinensis | | Descriptor: | Lipase, SULFATE ION | | Authors: | Zhang, M, Yu, X.W, Xu, Y, Huang, C.H, Guo, R.T. | | Deposit date: | 2018-06-06 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis by Which the N-Terminal Polypeptide Segment ofRhizopus chinensisLipase Regulates Its Substrate Binding Affinity.
Biochemistry, 58, 2019
|
|
5XK2
 
 | |
